FARSB stratifies prognosis and cold tumor microenvironment across different cancer types: an integrated single cell and bulk RNA sequencing analysis
ZHANG Ziran ,TAN Jiale ,YU Zihang ,LIU Chengdong ,WANG Jian ,WU Dehua ,BAI Xue
1Nanfang Hospital,Southern Medical University,Guangzhou 510515,China;2First School of Clinical Medicine,Southern Medical University,Guangzhou 510515,China
Abstract: Objective Immunotherapy has brought significant clinical benefits to a subset of patients,but has thus far been disappointing in the treatment of immunologically"cold"tumors.Existing biomarkers that can precisely identify these populations are insufficient.In this context,a potential cold tumor microenvironment(TME)marker FARSB was investigated to reveal its impact on TME and patients'response to immunotherapy across pan-cancer.Methods The expression levels and mutational landscape of FARSB in pan-cancer were investigated.Kaplan-Meier and univariate Cox regression analyses were applied to analyze the prognostic significance of FARSB.Pathways affected by FARSB were investigated by gene set enrichment and variation analysis.The relationship between FARSB expression and immune infiltration was examined using the TIMER2 and R packages.Single-cell RNA sequencing(scRNA-seq)data of several cancer types from GSE72056,GSE131907,GSE132465,GSE125449 and PMID32561858 were analyzed to validate the impact of FARSB on the TME.The predictive effect of FARSB on immunotherapy efficacy was explored in 3 immune checkpoint inhibitors (ICIs)-treated cohorts (PMID32472114,GSE176307,and Riaz2017).Results FARSB expression was significantly higher in 25 tumor tissues than in normal tissues and was associated with poor prognosis in almost all tumor types.FARSB expression exhibited a strong association with several DNA damage repair pathways and was significantly associated with TP53 mutation in lung adenocarcinoma (P<0.0001,OR=2.25). FARSB characterized a typical immune desert TME and correlated with impaired expression of chemokines and chemokines receptors.Large-scale scRNA-seq analysis confirmed the immunosuppressive role of FARSB and revealed that FARSB potentially shapes the cold TME by impeding intercellular interactions.In 3 ICI-treated cohorts,FARSB demonstrated predictive value for immunotherapy.Conclusion This study provides a pan-cancer landscape of the FARSB gene by integrated single-cell and bulk DNA sequencing analysis and elucidates its biological function to promote DNA damage repair and construct the immune desert TME,suggesting the potential value of FARSB as a novel marker for stratifying patients with poor immunotherapeutic benefits and"cold"TME.
Keywords:FARSB;immunotherapy;pan-cancer;DNAdamage repair;tumor microenvironment
INTRODUCTION
In recent years,the breakthrough concept of using the immune system to treat cancer has brought tremendous clinical benefits to cancer patients,but a large proportion of cancer patients still fail to respond to immune checkpoint inhibition (ICI) therapy[1-3],and even among those who respond,only a fraction of patients achieve durable remission[4,5].Understanding the mechanisms of immunotherapy resistance is therefore of great importance to expand the spectrum of tumor types and patients that may benefit from immunotherapy.With a better understanding of the multidimensional interactions between tumors,the immune system,and other systemic factors,researchers have proposed that the cold tumor microenvironment (TME,also known as immune desert) is one of the pivotal reasons for immunotherapy resistance[6].The causes and biomarkers of cold tumor immune microenvironment remain to be clarified,which drives a body of studies to identify and exploit immunological biomarkers or pathways that could be key to unlocking ICI therapy as a viable treatment option for immunologically cold tumors.
Phenylalanine tRNA synthetase (PheRS),a tetrameric protein consisting of two alpha (FARSA) and two beta(FARSB)subunits,typically functions to couple phenylalanine to homologous tRNAs required for the first step of protein synthesis[7,8].Patients with defective PheRS,encoded byFARSAandFARSB,were reported to display brain abnormalities,interstitial lung disease,and facial dysmorphism[9-11].In addition,Gao et al[12]found that the aminoacyl-tRNA biosynthesis pathway was upregulated in gastric cancer and thatFARSBplayed a key role in gastric cancer progression.Liu et al[13]revealed that the aberrant expression ofFARSBwas associated with proliferation of hepatocellular carcinoma cell and a poor survival outcome of the patients,suggesting the potential involvement ofFARSBin promoting tumor progression.These findings,however,are limited to specific cancer types,and whether they are applicable to other tumors is unclear.Beyond this,the pathways through whichFARSBaffects the tumor immune microenvironment and patient response to immunotherapy remains to be elucidated.
In this study,by analyzing large-scale single-cell and bulk sequencing data,we mapped for the first time the pan-cancer expression,prognosis,pro-DNA damage repair,and immunosuppression landscape ofFARSB,aiming to elucidate the impact ofFARSBon tumor response to immunotherapy.We further discovered thatFARSBserves as a biomarker for TME desertification by single-cell sequencing analysis.Our results provide a glimpse into the function of FARSB in pan-cancer and identifyFARSBas novel biomarker for TME classification and prediction of immunotherapeutic benefits.
METHODS
Data collection
The RNA expression and clinical data of pan-cancer samples were obtained from The Cancer Genome Atlas(TCGA) dataset (https://portal.gdc.cancer.gov/).The normal samples were selected from the Genotype-Tissue Expression (GETx) dataset.The combined cohort of TCGA and GTEx samples was downloaded from the UCSC Xena database (https://xenabrowser.net/datapages/)[14].The Human Protein Atlas (HPA,https://www.proteinatlas.org/) database[15]was used to explore the protein level ofFARSBin human tumor and normal tissues.
Prognostic analysis
Kaplan-Meier (KM) analysis was used to assess the overall survival (OS) of patients in the TCGA cohort.Univariate and multivariate COX regression analyses were performed to evaluate the significance ofFARSBin predicting OS in pan-cancer.
Gene Set Variation Analysis (GSVA) and Gene Set Enrichment Analyses(GSEA)
A "gmt" file (h.all.v7.4.symbols.gmt) containing 50 hallmark gene sets was downloaded from the Molecular Signature Database (MSigDB,https://www.gsea-msigdb.org/gsea/index.jsp) and used to calculate the normalized enrichment score (NES) and false discovery rate (FDR)for each biological process in each tumor.The Gene Ontology (GO),Kyoto Encyclopedia of Genes and Genomes (KEGG),and Reactome pathway were enrolled in GESA based on R-packages"limma","org.Hs.eg.db","clusterProfiler"and"enrichplot".
Genomic alterations analysis
The incidence of genomic alterations (including mutations,structural variations,amplifications,deep deletions,and multiple alterations) ofFARSBacross cancers was studied using the cBioPortal (http://www.cbioportal.org)[16,17].The rates of genomic alterations were visualized using bar graphs through cBioPortal WebTool.The mutation data in each tumor was downloaded from the UCSC XENA database,and the R package"maftools"was used to map gene mutations in high and lowFARSBexpression groups and to create forest plots showing the mutation frequency differences.
Immune cell infiltration
The correlation betweenFARSBexpression and ESTIMATE score,immune score,and stromal score was determined using ESTIMATE.We downloaded and analyzed the immune cell infiltration score of TCGA from TIMER2 database (http://timer.cistrome.org/)[18].For each TCGA tumor type,patients were divided into high and lowFARSBexpression groups based on the medianFARSBexpression level to compare the extent of immune cell infiltration.The relationship betweenFARSBand tumor mutational burden (TMB) and microsatellite instability (MSI) was analyzed in pan-cancer by Spearman correlation analysis.
Single-cell RNA-seq data acquisition and processing
Processed scRNA-seq matrix of tumor samples was downloaded under the accession name GSE72056(melanoma),GSE131907 (lung adenocarcinoma),GSE132465 (colorectal cancer) and GSE125449(hepatocellular carcinoma) from the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/).PMID32561858(breast carcinoma)was downloaded from a published study (PMID32561858)[19].The top 10 principal components and 2000 variable genes were involved in analyzing process using Seurat (version 4.2.0)[20].Dimensionality reduction was done by UMAP using the default settings of the RunUMAP function.Cell clusters in the resulting two-dimensional representation were annotated referring to the corresponding essays.
Correlation between TME and FARSB
To validate the role ofFARSBin scRNA datasets,the relation of immune cell and stromal cell fractions in TME with tumorFARSBexpression within samples were calculated.A scatter diagram and linear fitting were drawn to represent the results.
Ligand-receptor expression and cell interactions
To further assess the possible role of tumorFARSBin cell-to-cell communication,samples from GSE72056 and GSE131907 were both divided into high and lowFARSBexpression groups.CellChat(version 1.5.0)[21]was used to investigate the difference in ligand-receptor crosstalk in TME between high-and low-FARSBexpression groups.
Animal experiments
C57BL/6 mice (5-8 weeks old) of either sex with body weight of about 25 g were purchased from and maintained in the specific-pathogen-free facility of the Laboratory Animal Center of Southern Medical University(Guangzhou,China).A total of 10 mice were used in the study.C57BL/6 mice were injected subcutaneously with 2 × 106Lewis lung cancer cells (LLC1) in 100 μL PBS-matrigel mixture into the right leg.Two weeks after injection,the tumors were collected and processed for immunohistochemistry (IHC).The tumor tissues were fixed in 4% paraformaldehyde overnight,embedded in paraffin and sectioned.Antigen retrieval was performed by high-pressure heating with Tris-EDTA buffer(pH 9.0).H2O2(3%) was used to block endogenous peroxidase activity.After serum blocking,the sections were incubated overnight at 4 ℃with mouse FARSB(Proteintech,67924-1-Ig),CD8α (Abcam,ab217344),and F4/80(Abcam,ab300421)antibodies.After washing with PBST for 3 times,the sections were incubated with a secondary antibody for 1 h at room temperature.
Immunotherapy prediction analysis
To explore the relationship betweenFARSBand ICI therapy response,3 cohorts were used to validate the prediction ability ofFARSBfor immunotherapy response.PMID32472114 contains data of 181 patients with advanced clear cell renal cell carcinoma treated with PD-1 blockade[22].GSE176307 includes 90 patients with metastatic urothelial cancer treated with ICI,and Riaz2017 contains 22 patients with advanced melanoma treated with PD-1 blockade[23].Using the"Surv-Cutpoint"function of the"Survminer"R package,the patients were divided into low and highFARSBexpression groups based on the optimal cut-off value.SPSS(version 25)was used for multivariate COX regression analysis.TPM of CD274 andFARSBwas employed to calculate 95%confidential interval (95%CI) of the hazard ratios (HRs)in GSE91061 (57 melanoma patients) and GSE126044(16 non-small lung cancer patients).
Statistical Analyses
The Wilcoxon rank-sum test was performed to calculate the statistical significance and compareFARSBexpression levels between tumor and normal tissues.Pairedt-test was used to evaluate the statistical significance of the difference inFARSBexpression levels between tumor and adjacent tissues.Univariate Cox regression analysis and the Kaplan-Meier method were employed to assess the prognostic role ofFARSBexpression in each cancer.Spearman correlation analysis was performed to evaluate the statistical relationships betweenFARSBand other factors including immune cell infiltration levels,mismatch repair pathway,immune suppressive genes,and chemokine receptors.Finally,the chi-square test was used to compute the statistical significance to compare the proportions of ICI therapy responders and non-responders between the low and highFARSBgroups.All statistical analyses were performed using R 4.1.1,andP<0.05(two-tailed)was considered to indicate a statistically significant difference.
RESULTS
FARSB expression analysis in pan-cancer
We first assessedFARSBexpression in various types of cancers using the TCGA and GTEx datasets.As illustrated in Fig.1A,FARSBwas differentially expressed in 28 of 31 tumors compared to normal tissue,excluding mesothelioma (MESO) and uveal melanoma (UVM) for which normal tissue expression data were not available.Among them,25 tumor tissuesexhibited significantly higherFARSBexpression levels,including breast invasive carcinoma (BRCA),cholangiocarcinoma(CHOL),colon adenocarcinoma (COAD),glioblastoma multiforme(GBM),kidney renal papillary cell carcinoma(KIRC),lung adenocarcinoma (LUAD),and lung squamous cell carcinoma (LUSC).Only in kidney chromophobe (KICH),acute myeloid leukemia (LAML),and thyroid carcinoma (THCA),normal tissues showed higherFARSBexpression levels than tumors.TCGA paired expression analysis also confirmed that compared with the adjacent tissues,FARSB was expressed at high levels in BRCA,COAD,LUAD,and liver hepatocellular carcinoma (LIHC) (Fig.1B).Further,we explored the FARSB protein levels in the HPA database.Similarly,FARSB protein levels were lower in normal tissues than in tumor tissues (Fig.1C).These results suggest thatFARSBis significantly overexpressed at both mRNA and protein levels across the cancer types.
Prognostic role of FARSB across different cancers
We next investigated whetherFARSBaffected cancer patients' prognosis using univariate COX regression analysis.Survival metrics included OS,disease-specific survival (DSS),and progression-free interval (PFI).OS analysis showed thatFARSBacted as a risk factor in patients with LIHC(P<0.001,HR=2.160),adrenocortical carcinoma (ACC) (P<0.001,HR=3.367),LUAD (P<0.001,HR=1.599),BRCA (P=0.006,HR=1.539),bladder urothelial carcinoma (BLCA) (P=0.006,HR=1.419),KICH (P=0.017,HR=3.832),brain low-grade glioma (LGG) (P=0.020,HR=1.781),kidney renal papillary cell carcinoma (KIRP) (P=0.022,HR=2.263),pancreatic adenocarcinoma (PAAD) (P=0.033,HR=1.603),MESO (P=0.033,HR=1.719),and UVM (P=0.034,HR=2.347) (Fig.2A).Kaplan-Meier analysis,which stratified patients into high and lowFARSBexpression groups by the optimal cut-off value,also revealed that patients with lowFARSBlevels had favorable OS in most cancer types (Fig.2B).These findings indicate thatFARSBmay serve as a biomarker of unfavorable prognosis in pan-cancer.
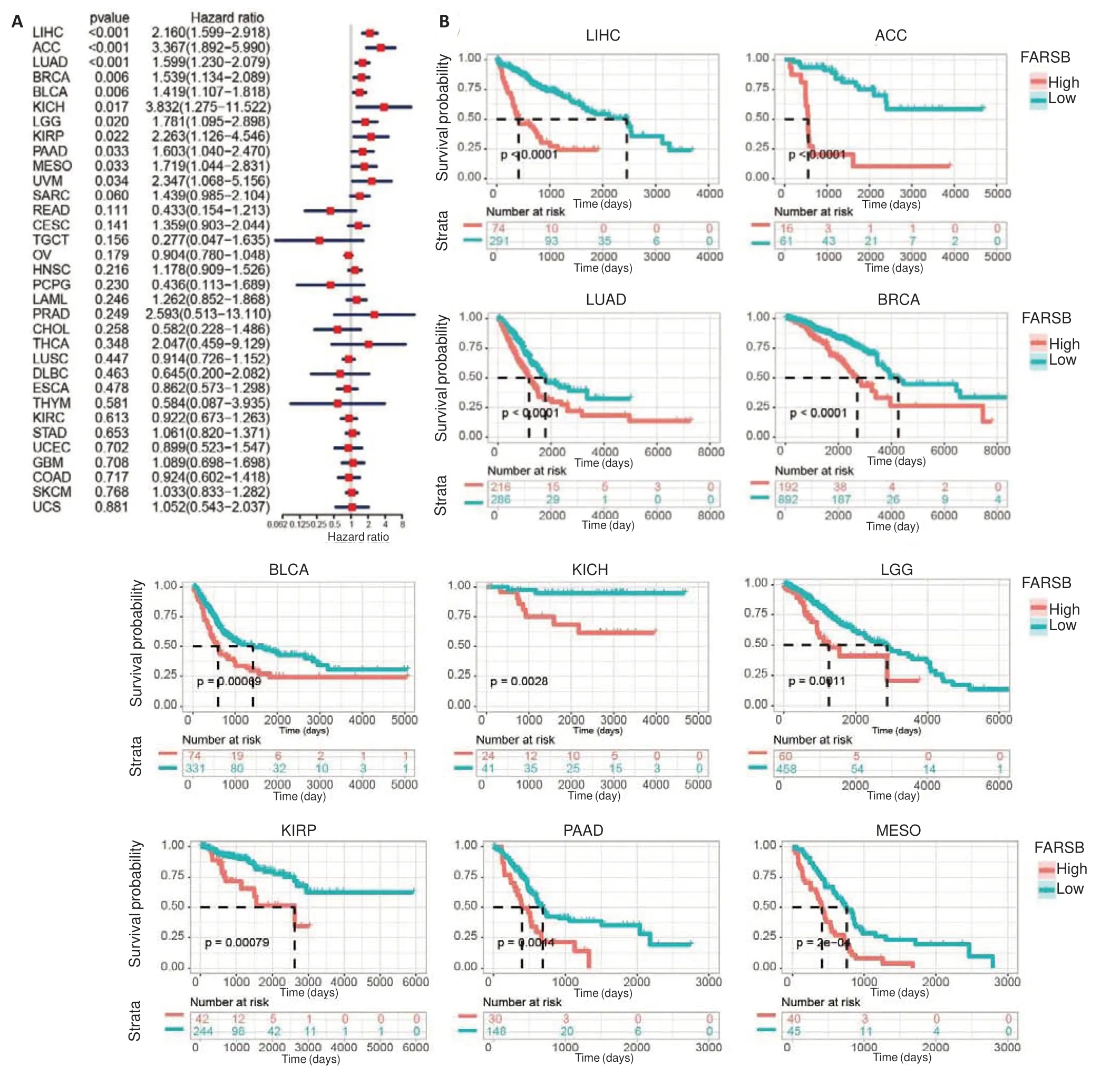
Fig.2 Survival analysis of FARSB in different cancer types from the TCGA database. A: Forest plot of survival analysis showing the impact of FARSB expression level on overall survival (OS) of patients with different cancer types.B:Prognostic value of FARSB for OS revealed by Kaplan-Meier analysis.
Biological significance of FARSB in different cancers
We next carried out GSVA and GSEA to explore the mechanisms underlying the association ofFARSBwith poor prognosis of the cancer patients.Among the 50 hallmark pathways,FARSBwas positively associated with proliferation-related processes such as MYC targets v1,MYC targets v2,and mTORC1 in almost all cancer types (Fig.3A),which is consistent with previously published data[12,13].Pathways associated with DNA damage repair such as G2M checkpoint and DNA repair were also enriched in patients with highFARSBexpressions.In addition,FARSBwas negatively correlated with myogenesis,xenobiotic metabolism,and coagulation in pan-cancer.GSEA based on GO and KEGG databases similarly revealed thatFARSBwas significantly correlated with DNA damage repair pathways in a wide range of tumor types (BRCA,KICH,LUAD,and LUSC;Fig.3B).Consistent with previous studies,FARSBaffected multiple DNA damage repair-related dimensions such as DNA damage checkpoints,double-strand break repair via homologous recombination (HR),recombination repair,non-recombination repair,mismatch repair (MMR),Fanconi anemia pathway[24],non-homologous end-joining(NHEJ),nucleotide excision repair (NER),and base excision repair (BER).We analyzed the difference in gene mutation frequency between high and lowFARSBexpression groups in LUAD and LUSC and found that the mutation frequency ofTP53,which promotes DNA damage repair[25],was significantly upregulated in LUAD patients with highFARSBexpression (OR=2.25),and KEAP1 gene mutation(OR=2.583)was more common in LUSC patients with high FARSB expression (Fig.3C,D).All these findings suggest thatFARSBplays a critical role in DNA damage repair.
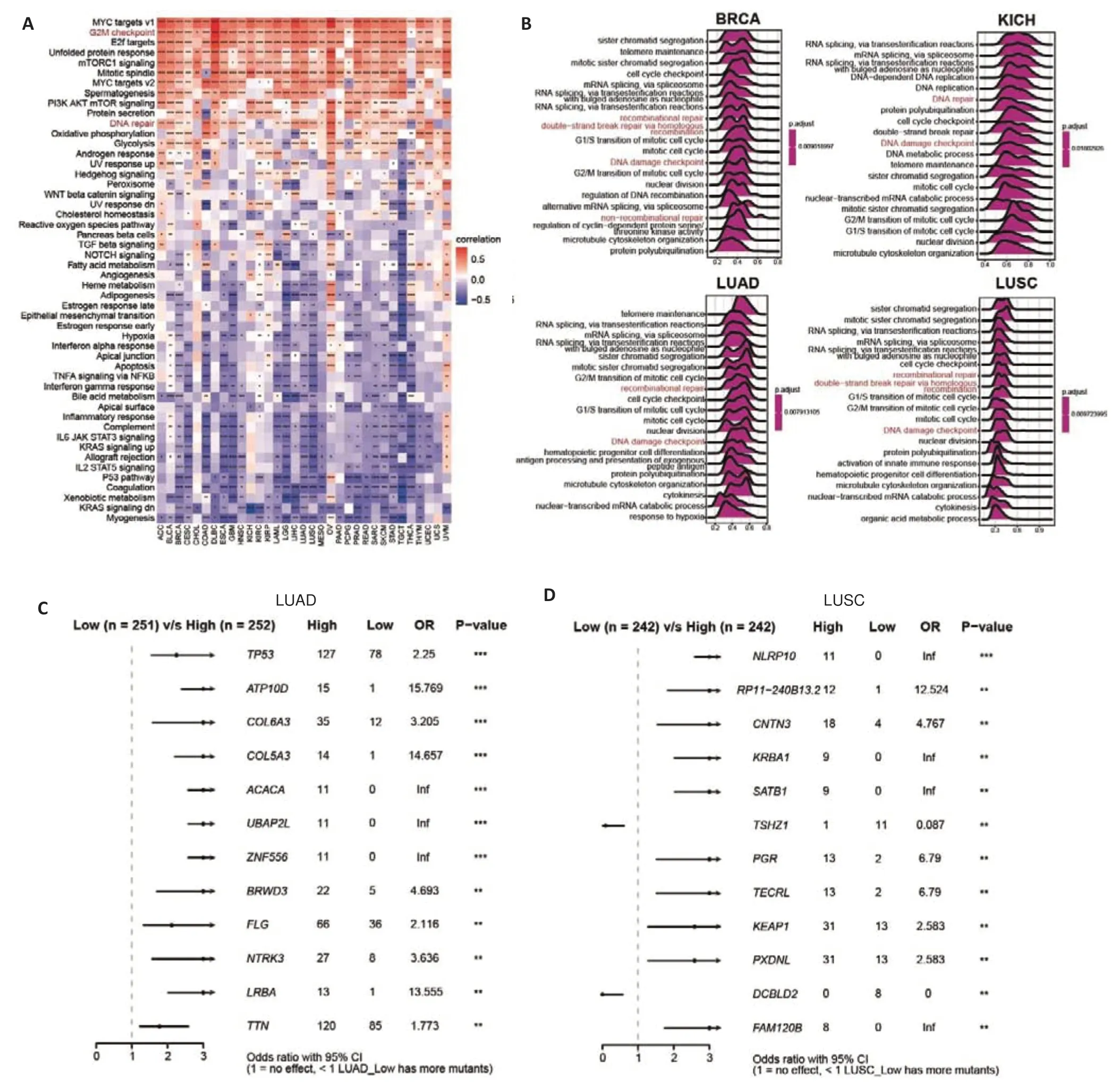
Fig.3 Pan-cancer functional analysis of FARSB levels.A:Functional enrichment pathways of FARSB analyzed using the GSVA algorithm.B:Functional enrichment pathways of FARSB analyzed using the GSEA algorithm based on the GO database in BRCA,KICH,LUAD,and LUSC.C,D:The difference of gene mutation frequency in FARSB high and low expression groups in LUAD(C)and LUSC(D).*P<0.05,**P<0.01,***P<0.001,****P<0.0001.
Immune desert TME identified by high expression of FARSB
Given thatFARSBis positively correlated with DNA damage repair-related pathways,which are related to tumor antigenicity and the patients'response to ICI[26],we performed a thorough analysis of the effect ofFARSBon TME.ESTIMATE analysis showed that in the majority of tumors,FARSBwas positively correlated with tumor purity while negatively with the immune score,stromal score,and ESTIMATE score(Fig.4A).Using the methods from a published study[27],we found that DNA damage response,MMR,NER,and BER pathways were significantly upregulated while antigen presentation machinery and CD8 effector T cells were markedly downregulated in patients with highFARSBexpression(Fig.4B,C).We also conducted a correlation analysis using immune cell infiltration data from the TIMER2 database (Fig.4D),and the results showed that the immune cells (including myeloid dendritic cells,macrophages,monocytes,NK cells,cells,CD4+T cells,and CD8+T cells) and stromal cells (including tumorassociated fibroblasts and endothelial cells) were significantly reduced in patients with highFARSBexpression.Moreover,FARSBsignificantly impaired the expression of chemokine receptors,which may affect cellular communication within the TME (Fig.4E).In addition,FARSBwas negatively correlated with immune checkpoint genes(such asCD274,HAVCR2,CTLA4,andLAG3) and positively with TGFBR1 in different cancers(Fig.4F).These results suggest thatFARSBnot only reduces the population of immune cells within the TME but also disrupts intercellular interactions,thus shaping a typical immune desert TME.
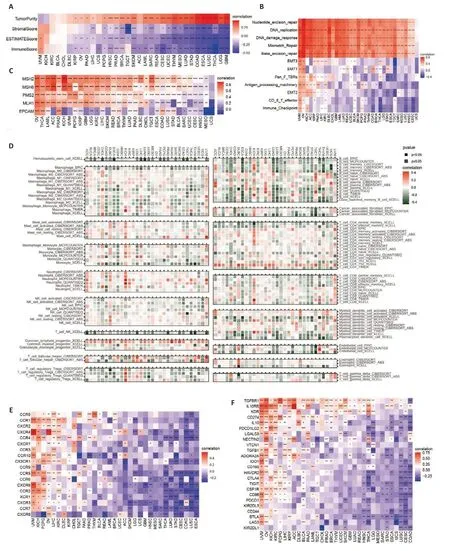
Fig.4 Immune aspects of FARSB in the tumor microenvironment. A: ESTIMATE analysis of FARSB in the majority of tumors,including tumor purity,immune score,stromal score,and ESTIMATE score. B: Relationship between FARSB expression and immune aspects using the methods from a published study. C: Relationship between FARSB expression and MMR genes. D: Correlation analysis of FARSB with immune cell infiltration using data from the TIMER2 database. E:Relationship between FARSB expression and chemokine receptor genes.F:Relationship between FARSB expression and immune checkpoint genes.*P<0.05,**P<0.01,***P<0.001,****P<0.0001.
Single-cell sequencing analysis validates the impact of FARSB on the TME
We next sought to investigate the impact ofFARSBon the TME in two single-cell sequencing datasets.We integrated and clustered single cells from patients in the melanoma cohort (GSE72056) and LUAD cohort(GSE131907) and found thatFARSBwas highly expressed predominantly in the tumor cells (Fig.5A,B).As consistent with the results shown in Fig.4C,the patients with highFARSBexpression had significantly reduced T cell and B cell infiltration in the TME(Fig.5C).Correlation analysis further confirmed that the expression level ofFARSBwas negatively correlated with the number of T and B cells and positively correlated with the number of tumor cells (Fig.5D).Similar results were observed in the LUAD cohort (Fig.5E-H) and in patients with breast carcinoma[19],colorectal cancer(GSE132465),and hepatocellular carcinoma (GSE125449) (Fig.5I-K).Immunohistochemistry of paired samples from C57BL/6 mice bearing Lewis lung tumors furtherly verified that tumors with highFARSBexpression displayed fewer infiltrating CD8+T cells and macrophages(Fig.5L).These findings corroborate the high expression ofFARSBin tumor tissues and the cold TME shaped by FARSB at the single-cell resolution.
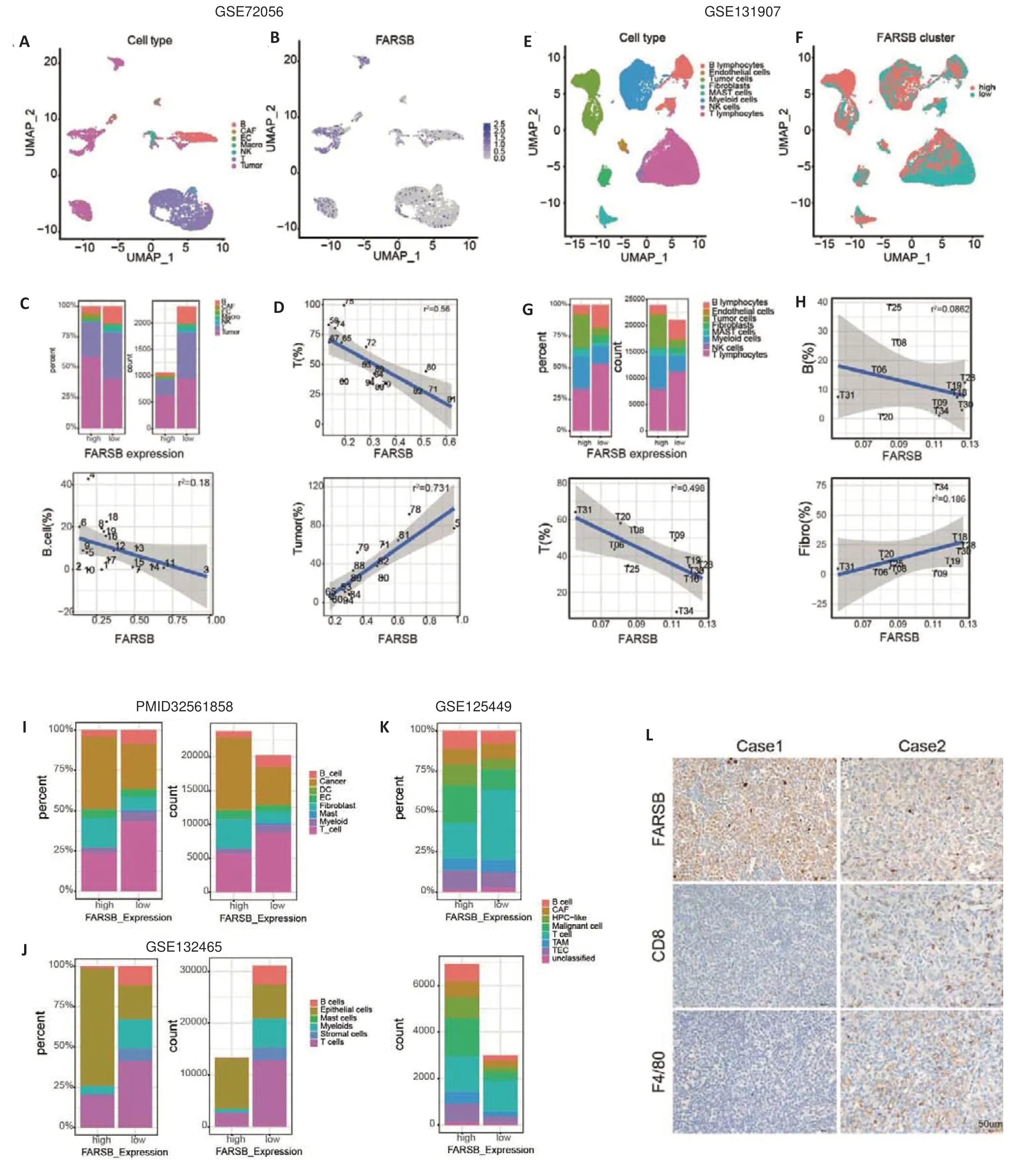
Fig.5 Single-cell sequencing and immunohistochemistry demonstrating the correlation between FARSB expression and the TME.Single cells from patients in the melanoma cohort(GSE72056)(A)and the LUAD cohort(GSE131907)(E)were clustered.B,F:FARSB expression in different cell clusters in the melanoma cohort and the LUAD cohort,respectively.C,G:Percentages and counts of infiltrating immune cells in high versus low FARSB expression groups in the melanoma cohort(C)and the LUAD cohort (G). D, H: Correlation analysis of the FARSB expression with the number of T cells,B cells,and tumor cells in the melanoma cohort (D) and the LUAD cohort (H). I-K: High and low expression of FARSB in relation to various immune cell infiltrations in breast carcinoma (I,PMID32561858),colorectal cancer (J,GSE132465),and hepatocellular carcinoma (K,GSE125449). L: Immunohistochemistry of FARSB,CD8 and F4/80 expression in paired samples from C57BL/6 mice bearing Lewis lung tumors(Scale bar:50 μm,×40).
Single-cell interaction analysis explains FARSBassociated cold TME formation
We further carried out an analysis of the cell-cell interactions within the TME in the above melanoma cohort.As shown in Fig.6A,in addition to the increased interactions between macrophages and other cells,the number and intensity of cell-cell interactions were significantly downregulated in melanoma patients with highFARSBexpression.More active crosstalk between T cells and other cells,both incoming and outgoing interactions,was also observed in patients with lowFARSBexpression (Fig.6B).We further explored the interactions of T cells with other cells and found that a highFARSBexpression impaired T cell contacts with the endothelial cells,macrophages,and NK cells (Fig.6C).Interaction pathway analysis suggested that T cell-endothelial cell contacts were enhanced in TME with lowFARSBexpression via ICAM1,which promoted T cell adhesion and infiltration (Fig.6D).Analysis of intercellular interactions in breast carcinoma,colorectal cancer,and hepatocellular carcinoma yielded similar results (Fig.6E-I).Taken together,FARSBpotentially inhibits the infiltration of T cells into the TME by interfering with their communication and thus constructing the immune desert TME.
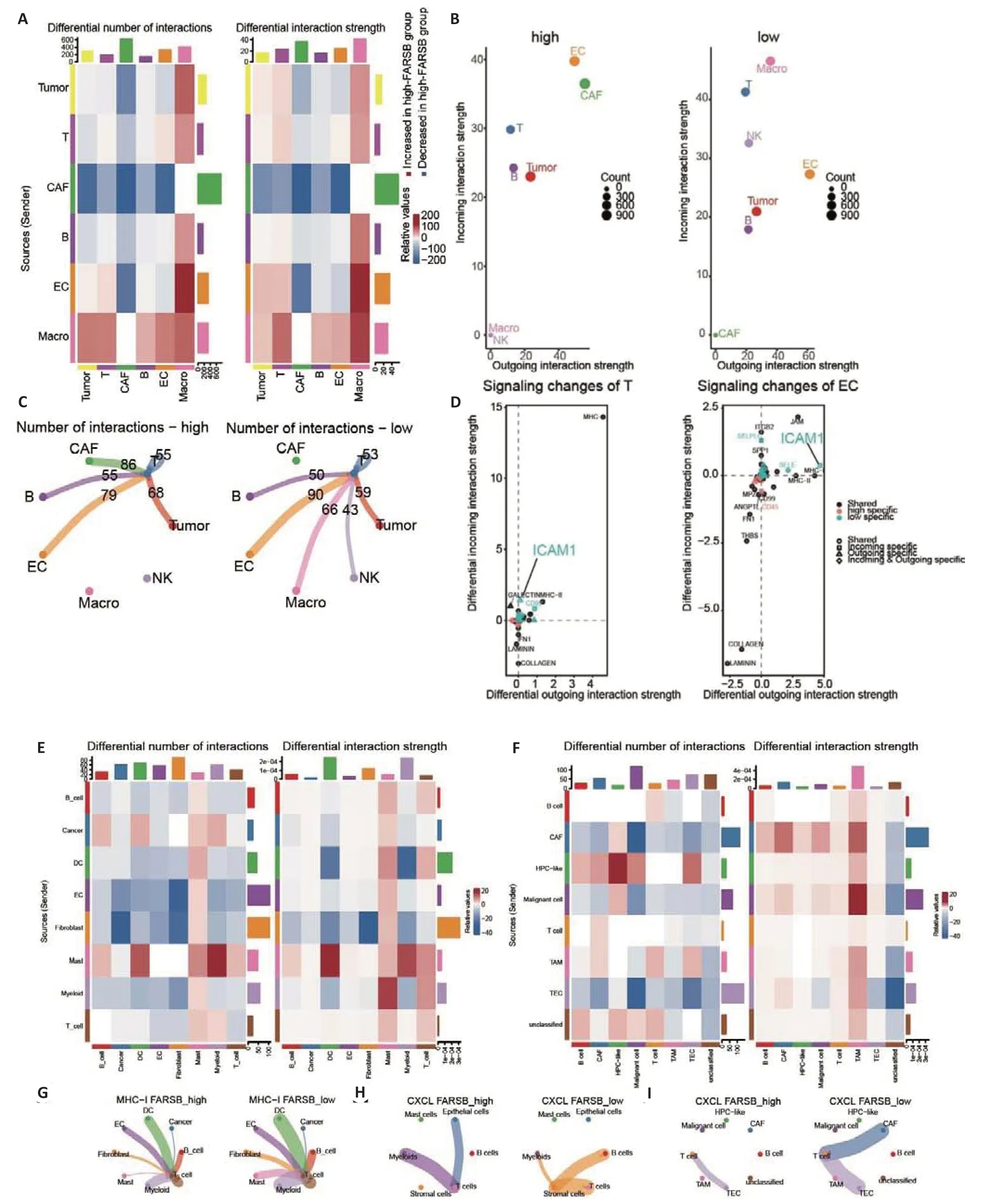
Fig.6 Single-cell interaction analysis between high and low FARSB expression groups in the melanoma cohort. A:Number and intensity of cell-cell interactions among those with high FARSB expression versus low FARSB expression.B:Incoming and outgoing interactions between T cells and other cells in tumors with high and low FARSB expression. C: Number of cell interactions in tumors with high and low FARSB expression. D: Interaction pathway analysis of T cell and endothelial cell contacts in tumors with high versus low FARSB expression.E-I:The number and intensity of cell-cell interactions among those with high FARSB expression vs low FARSB expression in breast carcinoma(E,G),colorectal cancer(H),and hepatocellular carcinoma(F,I).
Predictive value of FARSB for immune checkpoint inhibitors therapy.
Considering thatFARSBshapes immune desert in tumors,we explored whether its expression levels could discriminate immunotherapy efficacy in 3 cohorts treated with ICIs.As shown in Fig.7A,C and E,the patients with highFARSBexpression tended to have stable disease(SD) or progressive disease (PD) compared to those with low expression,while patients who responded to ICIs or reached partial remission (PR) or complete remission(CR) had lowerFARSBexpression levels.Additionally,the patients with lowFARSBexpression had significantly prolonged OS (Fig.7B,D) and progression-free survival(PFS) (Fig.7F) after receiving immunotherapy.We further performed a multivariate COX regression analysis in two additional cohorts receiving immunotherapy and found thatFARSBwas an independent risk factor for OS in the melanoma cohort (Fig.7G;HR=1.096,P=0.002,GSE91061)and for PFS in the non-small cell lung cancer cohort (Fig.7H;HR=1.002,P=0.003,GSE126044),further illustrating the impact ofFARSBon immunotherapy.These results imply that a high expression ofFARSBmay serve as a predictive biomarker for poor response to immunotherapy.
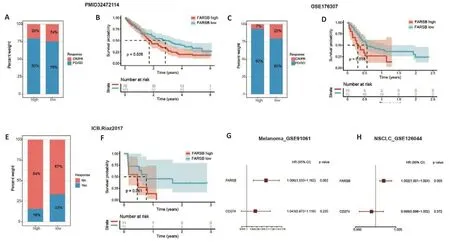
Fig.7 Immunotherapy efficacy prediction and sensitive drug analysis for FARSB indifferent cancers.A-D:Immunotherapy response(A,C)and overall survival(B,D)of ICI-treated cohorts(PMID32472114 and GSE176307)with high and low FARSB expression.E,F:Response to immunotherapy(E)and PFS(F)of patients with FARSB high and low expression in ICI-treated cohort(Riaz2017).G,H:Multivariate Cox regression analysis in in the melanoma cohort(G,GSE91061)and in the non-small cell lung cancer cohort(H,GSE126044).
DISCUSSION
By analyzing large-scale single-cell and bulk sequencing data,we comprehensively examined the expression,prognostic value,and biological significance ofFARSBand its impact on the TME in a pan-cancer dataset.The results showed thatFARSBwas significantly upregulated in multiple tumors as compared with normal and para-cancerous tissues,and was associated with worse OS,DSS,and PFI.GEVA and GESA analyses revealed thatFARSBpromoted DNA damage repair in multiple dimensions.TME analysis showed a differential reduction of the stromal cells and infiltrating immune cells in the TME with highFARSBexpression.Singlecell sequencing analysis of melanoma and LUAD cohorts confirmed thatFARSBimpeded T and B cell infiltration and impaired cell-cell interactions in the TME,which may contribute to the formation of immunologically cold tumors.Finally,we explored the feasibility ofFARSBas a predictive marker for treatment responses in 3 immunotherapy cohorts,and the results showed that a high expression ofFARSBwas related to poor immunotherapy efficacy.
In a given malignancy,the genetic status of the tumor is one of the main determinants of response to ICI[26].During tumor development,the tumor cells acquire several mutations and produce mutated proteins and peptides,which can serve as neoantigens,different from autoantigens.In many tumors,the neoantigens are not protected by self-tolerance mechanisms and are therefore immunogenic[28].In addition,genetic abnormalities in the tumor cells can cause abnormal expressions or localization of autoantigens.The neoantigens and abnormal autoantigens expressed in the tumor tissue can attract T cells to eliminate the tumor cells,which enhances anti-tumor immune response induced by ICIs[26].Therefore,TMB and MSI have been used as criteria for determining tumor antigenicity and assessing response or resistance to ICIs[29-32].In many scenarios,in order to maintain the structural integrity of the genome,cells are capable of repairing DNA damage caused by exogenous factors using a variety of mechanisms,including BER,NER,MMR,HR repair,and NHEJ repair[33].In this study,we found thatFARSBwas associated with multiple DNA damage repair pathways,suggesting a potential role ofFARSBin repairing intracellular DNA damage to maintain genomic stability and reduce TMB levels.However,the correlation betweenFARSBand TMB was weak according to the results of correlation analysis.One possible explanation is that several pathways and multiple genes are involved in the DNA damage repair mechanisms,in whichFARSBmay not play the role of a central player asBRCA1/2does in homologous recombination repair[34-36].Very likely the expression level ofFARSBonly has a marginal effect on TMB;this is why a large number of DNA damage repair-related genes have to be included for predicting immunotherapy efficacy[37].In addition,the contribution of different DNA damage repair pathways to TMB augmentation varies across different cancer types: in non-small cell lung carcinoma(NSCLC),the BER pathway has significantly greater contribution to TMB than the HR or checkpoint pathways[38].
Immune desert tumors lack T cells and mainly exhibit neuroendocrine features or contain highly proliferative tumor cells,as in pediatric malignancies,hormone receptor-positive breast cancer,prostate cancer,glioblastoma,and small cell lung carcinoma (SCLC).These cancer types,except for SCLC,generally show poor responses to immune checkpoint inhibitors[6,39-41].The potential drivers of the immune desert phenotype are complex and not fully understood[6].Our data suggest thatFARSBshapes the typical immune desert TME,and the mechanisms behind it remain to be further elucidated.Possible explanations are thatFARSBpromotes DNA damage repair to reduce the production of tumor neoantigens,which in turn damages T cell infiltration[26];or that the pro-DNA damage repair effect ofFARSBleads to a decrease in cytoplasmic DNA,thereby attenuating the activation of the intrinsic immune system by the STING pathway[42].In addition,FARSBinterference with intercellular interactions within the TME,which results in inactive cell crosstalk,may also be one of the underlying mechanisms.
Immunotherapy is rapidly reshaping the landscape of clinical oncology.Although ICIs induce durable responses in many patients,there is wide variation in their efficacy both within and across tumor types[42].A major challenge lies in the identification of predictive biomarkers that can guide ICIs.The current predictors mainly include genetic biomarkers and immune biomarkers.Several studies supported the value of TMB,MSI,and mismatch repair deficiency as biomarkers to predict response to ICI[30,43,44].In multiple cancer types,the pretreatment levels of tumor-infiltrating lymphocytes(TILs) and immune checkpoint molecules are correlated with anti-PD-1 and anti-PD-L1 antibody responses[45-48].Although the value of these biomarkers has been demonstrated in specific cancer types,given the complexity of tumor immune response and ICI resistance mechanisms,these biomarkers alone can not meet the prognostic and predictive needs for patients treated with ICI[26],and better predictive strategies are needed.In our case,FARSBdemonstrates a favorable predictive efficacy,and when combined with the aforementioned biomarkers,it is expected to allow more accurate identification of the patients benefiting from immunotherapy.Nevertheless,our systematic analysis of FARSB has some limitations.We examined the correlation ofFARSBwith immunotherapy efficacy based on data from patients receiving immunotherapy as a second-line treatment,while the data from first-line treatment cohorts are not available.Therefore,prospective clinical trials are needed to elucidate the ability ofFARSBto predict the efficacy of ICIs.
In conclusion,FARSBhas pleiotropic effects in cancers and plays versatile roles in promoting tumor proliferation,facilitating DNA damage repair,and shaping the immune desert TME.Our data may help to better understand the formation of the immune desert TME and support the value ofFARSBas a potential biomarker for immunotherapy efficacy prediction.Targeting theFARSB-dependent signaling might become a promising therapeutic strategy for cancer immunotherapy.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon request.
 南方醫(yī)科大學(xué)學(xué)報(bào)2023年5期
南方醫(yī)科大學(xué)學(xué)報(bào)2023年5期
- 南方醫(yī)科大學(xué)學(xué)報(bào)的其它文章
- EHHADH是肝細(xì)胞癌脂肪酸代謝通路的關(guān)鍵基因:基于轉(zhuǎn)錄組分析
- 左歸降糖解郁方促進(jìn)糖尿病并發(fā)抑郁癥大鼠的海馬齒狀回神經(jīng)干細(xì)胞的自我更新并激活Shh信號(hào)通路
- 蘆薈苷可抑制胃癌細(xì)胞的增殖和遷移:基于下調(diào)STAT3/HMGB1信號(hào)通路
- RITA 體外選擇性抑制BAP1缺失的皮膚黑色素瘤細(xì)胞的生長(zhǎng)
- 激活小鼠ZI區(qū)GABA 能神經(jīng)元可促進(jìn)七氟醚和丙泊酚的麻醉誘導(dǎo)而對(duì)麻醉維持及覺醒無影響
- S100A10可促進(jìn)肺腺癌細(xì)胞的增殖和侵襲:基于激活A(yù)ktmTOR信號(hào)通路
