Endosymbiotic pathogen-inhibitory gut bacteria in three Indian Major Carps under polyculture system: A step toward making a probiotics consortium
Koushik Ghosh, Anjn Mukherjee, Dipnjn Dutt, Sudeshn Bnerjee,Ev Mrie Breines, Ellinor Hreide, Einr Ring?
aAquaculture Laboratory, Department of Zoology, The University of Burdwan, Burdwan, 713104, India
bDepartment of Arctic and Marine Biology, Faculty of Bioscience, fisheries and Economics, UiT, The Arctic University of Norway, Breivika, 9037, Norway
cNorwegian College of fishery Science, Faculty of Biosciences, fisheries and Economics, UiT, The Arctic University of Norway, Troms?, 9037, Norway
ABSTRACT
Keywords:
Carp
Bacillus
Stenotrophomonas
Antagonism
Exoenzymes
16S rRNA
1.Introduction
Aquaculture is an important food sector for a growing global human population and has rapidly developed due to intensified culture methods(FAO, 2017). The major producer countries in farmed fish are China,India, Vietnam, Bangladesh and Egypt (FAO, 2016, p. 200). In India,Indian major carps (IMCs), i.e., rohu (Labeo rohita), catla (Catla catla)and mrigal (Cirrhinus mrigala) accounting for almost 87% of the total freshwater fish production, and these fish species represent different trophic levels and form the most important component of the carp polyculture system (ICLARM, 2001; Paul & Giri, 2015). Polyculture of carps representing different ecological niche is a traditional method for optimum utilization of trophic resources in culture ponds (Billard &Berni, 2004). However, extension, diversification and intensification of aquaculture have increased the occurrence of disease outbreaks during the past decades (Mukherjee et al., 2017), and bacteria are the most common among the pathogens in cultured fish that cause mass mortality in freshwater aquaculture (Giri et al., 2011; Swain, Behura, Dash, &Nayak, 2007). Suggested correlations between modulation in the gut microbiota with physiology and disease have received increased attention of the scientific community leading to detailed investigations on the microbial diversity in fish (Ghanbari, Kneifel, & Domig, 2015; Hoseinifar, Sun, Wang, & Zhou, 2018). A comprehensive investigation of the gut-associated microbiota of the host might shed light on the “normal”bacterial community that could help to maintain fish health under polyculture. Although several studies have condemned culture-dependent methods as they detect only a small fraction of the microbial communities (Gajardo et al., 2016; Ghanbari et al., 2015; Kim,Brunt, & Austin, 2007; Larsen, Tao, Bullard, & Arias, 2013), de Bruijn,Liu, Wiegertjes, and Raaijmakers (2018) stated in their review that classic culture-dependent techniques are required to validate the potential of probiotic bacteria.
The gut is one of the major infection routes in fish because they are always in intimate contact to their environment, water, and are permanently exposed to bacteria including pathogens. The gut microbiota of fish plays an important role in mediating and stimulating host gastrointestinal development, aiding digestive function, maintaining mucosal tolerance, stimulating the host immunoresponse and providing a level of protection against gastric infections (Clements, Angert,Montgomery, & Choat, 2014; Montalban-Arques et al., 2015; Rawls,Mahowald, Goodman, Trent, & Gordon, 2007; Rawls, Mahowald, Ley, &Gordon, 2006; Ring? et al., 2016). However, indiscriminate use of chemical additives and antibiotics as preventative measure towards diseases has resulted in antimicrobial resistance among pathogenic bacteria, alteration in the gut microbial community and degraded environmental conditions (Cabello, 2006; Romero, Feijoo’, & Navarrete,2012; Ring? et al., 2016). Consequently, the scientific community has searched for alternatives, for example the probiotics. At present, there is a growing interest on the application beneficial microorganisms as probiotics to reduce the incidence of fish diseases by inhibiting the growth of pathogenic microorganisms (Balc′azar et al., 2006; Kesarcodi-Watson, Kaspar, Lategan, & Gibson, 2008; Mukherjee, Chandra, &Ghosh, 2019a; Munir, Hashim, Nor, & Marsh, 2018; Nandi, Banerjee,Dan, Ghosh, & Ray, 2018) and to improve the nutrient utilization(Mukherjee et al., 2019a; Verschuere, Rombaut, Sorgeloos, & Verstraete, 2000).
In their review devoted to probiotic and prebiotics for salmonids,Merrifield et al. (2010) extended a list of criteria for potential probionts,in which some were considered as essential while others as merely favorable. Some of the essential characteristics are: (1) must not be pathogenic to the host species, (2) must be resistant to bile salts and (3)low pH. Among the favorable criteria, functionally pertinent to pursue are: (4) should be able to adhere to and/or grow well within intestinal mucus, (5) must be free of plasmid-encoded antibiotic resistance genes,(6) should exhibit antagonistic properties towards one or more key pathogens and (7) should produce relevant extracellular digestive and/or degradation enzymes (e.g. cellulase, if the diet is rich in plant ingredients). The main strategy of using probiotics is to isolate intestinal bacteria with favorable properties from mature animals and include them in the feed for immature animals of the same species (Gildberg,Mikkelsen, Sandaker, & Ringo, 1997; Hoseinifar et al., 2018; Van Doan et al., 2018). However, unlike monoculture of salmon, tilapia, rainbow trout or sea bass, the aquaculture in India is typically practiced as composite culture of carps. Consequently, application of probiotics should cross the source species barrier to ensure overall health benefit to the fish species under composite culture practice. Therefore, multi-strain and multi-species probiotics should be developed from different fish species to cover wide angel benefits under composite culture conditions.
In a recent review, Lescak and Milligan (2017) put forward the controversial statement that teleosts should be used as model organisms to understand host-microbe interactions, and that the adherent(autochthonous symbiotic) microbiota should be investigated. The aim of the present study was to investigate autochthonous endosymbiotic gut bacteria isolated from three Indian major carp species, in order to isolate potential probiotics based on; functional characterization (antibacterial activity, enzymatic production), stability within the gut micro-environment (growth in mucus, tolerance to bile juice), bio-safety(hemolytic activity, antibiotic susceptibility, in vivo validation through intra-peritoneal injection), and to identify the bacteria by 16S rRNA partial gene sequence analyses.
2.Material and methods
2.1.Sample collection and isolation of autochthonous gut bacteria
Healthy fish with no external wound or sore were collected from three different polyculture ponds in and around Burdwan (2314N,8739E), West Bengal, India. Specimens were collected and handled following the approved guidelines of the Institutional Ethical Committee. However, approval of the committee was not required as farmed specimens were used. Three Indian major carps viz., rohu (Labeo rohita);catla (Catla catla); and mrigal (Cirrhinus mrigala) were used in the present study. Five fish specimens (average weight: 225 ±10.2 g; length 29.1 ±2.64 cm) of each species were collected from each of the three composite culture ponds. Pooled sample of each species collected from a particular pond served as a replicate, and thus the study comprised three replicates for each species. The specimens were starved for 24 h to isolate autochthonous endosymbiotic intestinal bacteria and to eliminate most of the allochthonous bacteria associated with digesta (Ghosh,Roy, Kar, & Ring?, 2010; Mukherjee et al., 2017). After starvation, fish were anaesthetized with MS-222 (tricaine methanesulfonate;Sigma-Aldrich Corp., USA) before sacrifice. The gastrointestinal (GI)tracts were divided into proximal (PI) and distal (DI) segments and the gut samples were processed for isolation of culturable autochthonous gut bacteria by the methods described previously (Mandal & Ghosh,2013; Mukherjee et al., 2016; Mukherjee & Ghosh, 2016). Gut segments were homogenized, serially diluted and spread on soybean casein digest medium (tryptone soya agar, TSA; HiMedia). Following incubation (48 h, 30C), distinct colonies were randomly isolated, cultured on TSA plates and pure cultures were preserved (4C) for further studies.
2.2.Antimicrobial activity assay
Antibacterial activity of the isolated gut bacteria was tested towards seven fish pathogenic strains by the ‘double-layer’ method of Dopazo et al. (1988). The pathogenic strains, Aeromonas hydrophila MTCC-1739(AH), Aeromonas salmonicida MTCC-1945 (AS), Aeromonas sobria MTCC-3613 (ASo), Pseudomonas fluorescens MTCC-103 (PF), Pseudomonas putida MTCC-1072 (PP) and Bacillus mycoides MTCC-7538 (BM)were obtained from the Microbial Type Culture Collection, Chandigarh,India, while, Aeromonas veronii KT737240 (AV) was isolated from a diseased catla (Mukherjee & Ghosh, 2016). Growth inhibition of the pathogenic strains was determined as halo zones and presented as inhibition scores: 0 (0-5 mm), 1 (low, 6-10 mm), 2 (moderate, 11-20 mm), 3 (high, 21-25 mm), and 4 (very high, ≥26 mm). The most promising antagonistic bacteria were primarily selected based on cumulative inhibition scores ≥13.
2.3.Molecular identification and phylogenetic analysis
All antagonistic bacteria were analysed by 16S rRNA partial gene sequences as described elsewhere (Mukherjee & Ghosh, 2016; Ring?,Sperstad, Myklebust, Mayhew, & Olsen, 2006). Universal primers, 27f(5-AGAGTTTGATCCTGGCTCAG-3) and 1492r (5-GGTTACCTTGT TACGACTT-3) were employed to amplify the gene encoding 16S rRNA.Amplified products were sent to the commercial house for Sanger sequencing using automated DNA sequencer (Applied Biosystems, Inc.,Foster City, CA, USA). Sequenced data were edited (BioEdit Sequence Alignment Editor; Version 7.2.5), the closest known (type strain)alignment identities were retrieved from National Centre for Biotechnology Information (NCBI) GenBank, and deposited to the NCBI Gen-Bank to obtain accession numbers.
2.4.Enzymatic activity assay
The selected antagonistic strains were further screened for production of extracellular digestive (amylase, protease, lipase) and antinutritional degrading (cellulase, xylanase, phytase) enzymes. The bacteria strains grown in selective broth media were analysed for production of the enzymes. Quantitative determination of amylase (Bernfeld,1955), protease (Walter, 1984, pp. 270-277), lipase (Bier, 1955),cellulase (Denison & Koehn, 1977), xylanase (Bailey, Biely, & Poutanen,1992) and phytase (Yanke, Selinger, & Cheng, 1999) activities were carried out following standard methodologies and expressed as unit activity (U).
2.5.Stability in gut micro-environment: growth in mucus and tolerance to bile juice
A description relating to growth potential of bacteria in fish mucus and bile tolerance has been depicted elsewhere (Balc′azar et al., 2008;Mukherjee et al., 2017; Mukherjee & Ghosh, 2016; Nikoskelainen, Salminen, Bylund, & Ouwehand, 2001). Mucus from intestine and skin of live carp specimens (average weight 145.45 ± 8.7g; length 16.8 ± 1.27 cm) was collected and processed separately following the methods described by Mukherjee and Ghosh (2016) and Ross, Firth, Wang, Burka,and Johnson (2000), respectively. Protein concentration of the mucus was determined (Lowry, Rosenbrough, Fair, & Randall, 1951) and adjusted to 1 mg mL. Mucus samples were filter sterilized (0.8 and 0.22 μm porosity; HiMedia, Mumbai, India) and inoculated with the selected strains (30C, 24 h) to confirm their growth potential in fish mucus. Crude bile juice (pH 5.7) was collected by puncturing gall bladder taken out from live specimens (IMCs),filter sterilized and stored at -20C for further use. Sterile PBS supplemented with 20% (v/v) fish bile juice was inoculated with the selected bacteria, incubated (30C,1.5 h) and viable counts were determined by spreading on TSA media plates.
2.6.Bio-safety assay
2.6.1.Hemolytic activity
Selected strains were investigated for hemolytic activity to determine their pathogenic potential (Nurhidayu, Ina-Salwany, Mohd-Daud,& Harmin, 2012). The assay was performed by streaking the bacteria cultures onto plates containing Columbia blood agar base (HiMedia,India) supplemented with goat blood (5%) and incubated at 30C for 24 h. Appearance of hemolytic zones around the colonies were noticed and classified as: α (greenish halo), β (clear halo) or γ (no halo) hemolysis based on lysis of the red blood cells in the media around and under the colonies.
2.6.2.Determination of antibiotic susceptibility
Antibiotic susceptibility of the selected strains was determined on TSA plates with susceptibility test discs (HiMedia, India) following discdiffusion methodand zones around discs were measured. The studied antibiotics (Ampicillin, Amoxicillin, Azithromycin, Chloramphenicol,Clindamycin, Erythromycin, Gentamicin, Kanamycin, Neomycin,Novobiocin, PenicillinG, Streptomycin, Tetracycline, Vancomycin) were used at prescribed doses and sensitivity was determined following the recommendation of National Committee for Clinical Laboratory Standards (NCCLS, 2012).
2.6.3.Small-scale in vivo validation
In vivo bio-safety evaluation for each of the γ-hemolytic bacteria was carried out separately as described by Mukherjee and Ghosh (2016) and Mukherjee et al. (2017). Brie fly, experimental fish (rohu, 15.6 ±1.2 g)were given intra-peritoneal (IP) injection (1.0 mL) of a selected bacterium (10cells/mL, in sterile 0.9% saline) and observed for 4 weeks for development of any external pathological symptoms (loss of scale or mucus, hemorrhage, lesion). Control fish were injected with sterile 0.9% saline (Mesalhy, Abd-El-Rahman, John, & Mohamed, 2008).
2.7.Statistical analysis
Results on exo-enzyme producing ability, growth potential in fish mucus and bile tolerance were presented as mean ±standard error (SE).Data on exo-enzyme producing ability was subjected to analysis of variance following Zar (1999) using SPSS version 17 (Kinnear & Gray,2009), and differences between means were tested by Tukey’s range test(P ≤0.05).
3.Results
3.1. fish species, bacterial isolates and antimicrobial activity
Totally, 1216 strains were randomly isolated from the three IMCs, of which, 545 strains were isolated from PI and 671 strains from DI.Amongst them, 47 strains from PI (8.62%) and 58 strains from DI(8.64%) exhibited antagonistic activity against at least one of the pathogens evaluated. Total number of isolates from PI and DI regions and antagonistic isolates from respective portions with reference to each fish species are presented in Fig. 1. While demonstrating pathogeninhibition by the isolated strains, 53 strains that revealed antagonism against ≥4 pathogenic strains and acquired cumulate inhibition score of≥10 (with respect to halo zone) are presented in Table 1. Of these, 27 promising pathogen inhibitory strains were primarily selected from rohu(16), catla (4) and mrigal (7) on the basis of a cumulative inhibition score of ≥13 and were further characterized to validate their potential probiotic attributes.
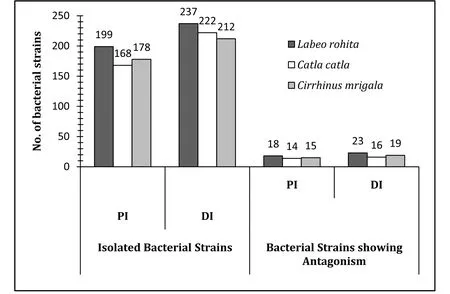
Fig. 1.Bacteria strains isolated from the proximal (PI) and distal (DI) regions of the gut in Indian major carps.
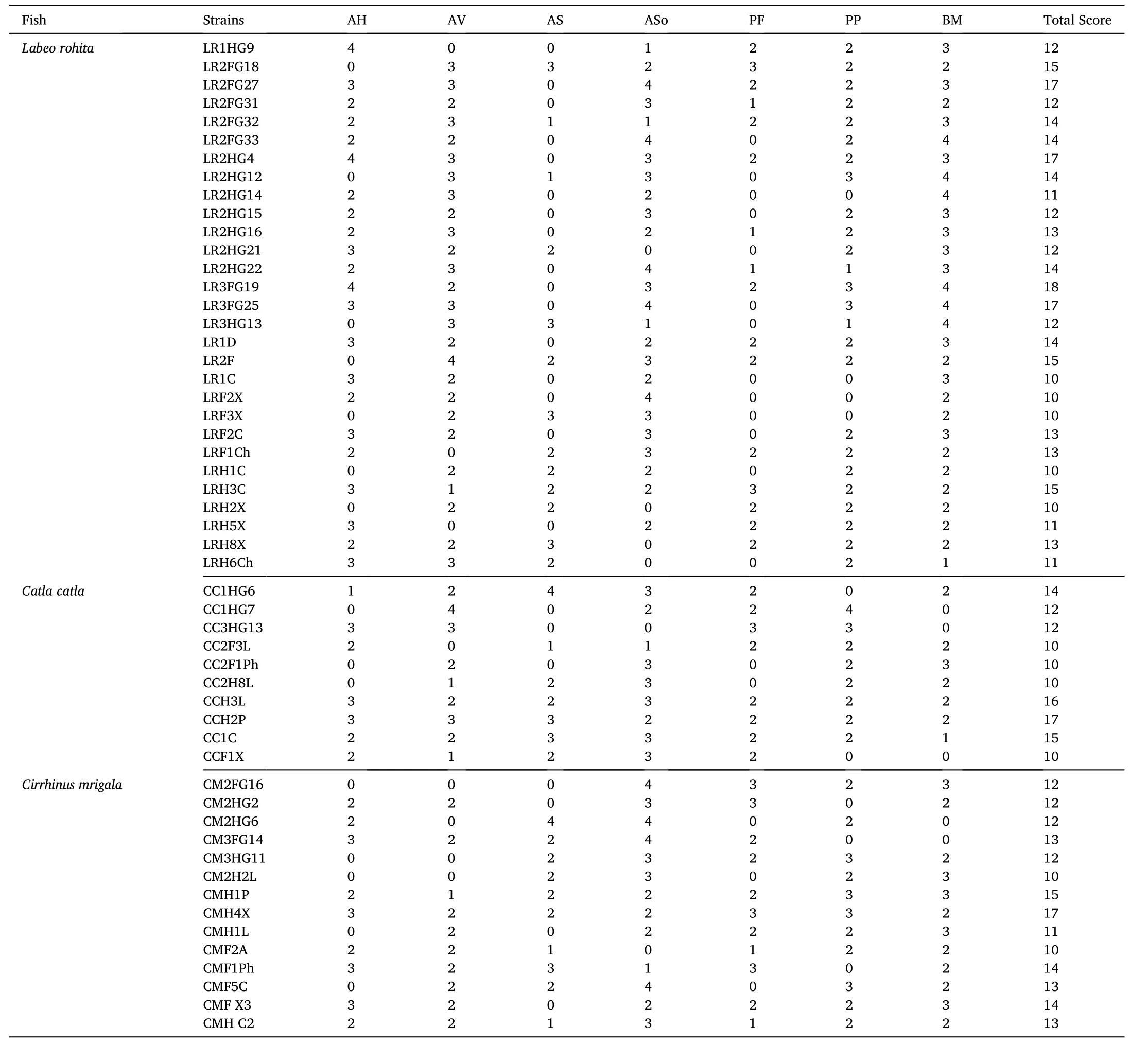
Table 1Determination of antagonism (double layer method) by the isolated gut bacteria against fish pathogens. Zones of inhibition (halo diameter) were presented as scores?.
3.2.Molecular identification and phylogenetic analysis
Identity of the pathogen-inhibitory bacteria as evidenced through nucleotide homology and 16S rRNA partial gene sequence analyses are depicted in Table 2. Out of the 105 pathogen-inhibitory gut isolates, 99 strains (94.29%) belonged to the genusBacillus
(similarity between 94 and 100%), while the other isolates were represented byPseudomonas fluorescens
(similarity =98%), Micrococcus aloeverae
(similarity =99%),Micrococcus yunnanensis
(similarity =89%), Stenotrophomonas pavanii
(similarity =99%), Lactococcus lactis
(similarity =97%) andStaphylococcus capitis
(similarity =99%). Bacteria identified asBacillus licheniformis
were most common (28%) among the pathogen-inhibitory bacteria, followed byB. safensis
(17%) andB. aerius
(12%). Diversity of the pathogen-inhibitory bacteria at species level as appeared through molecular identification of the isolated autochthonous pathogeninhibitory bacteria is presented in Fig. 2.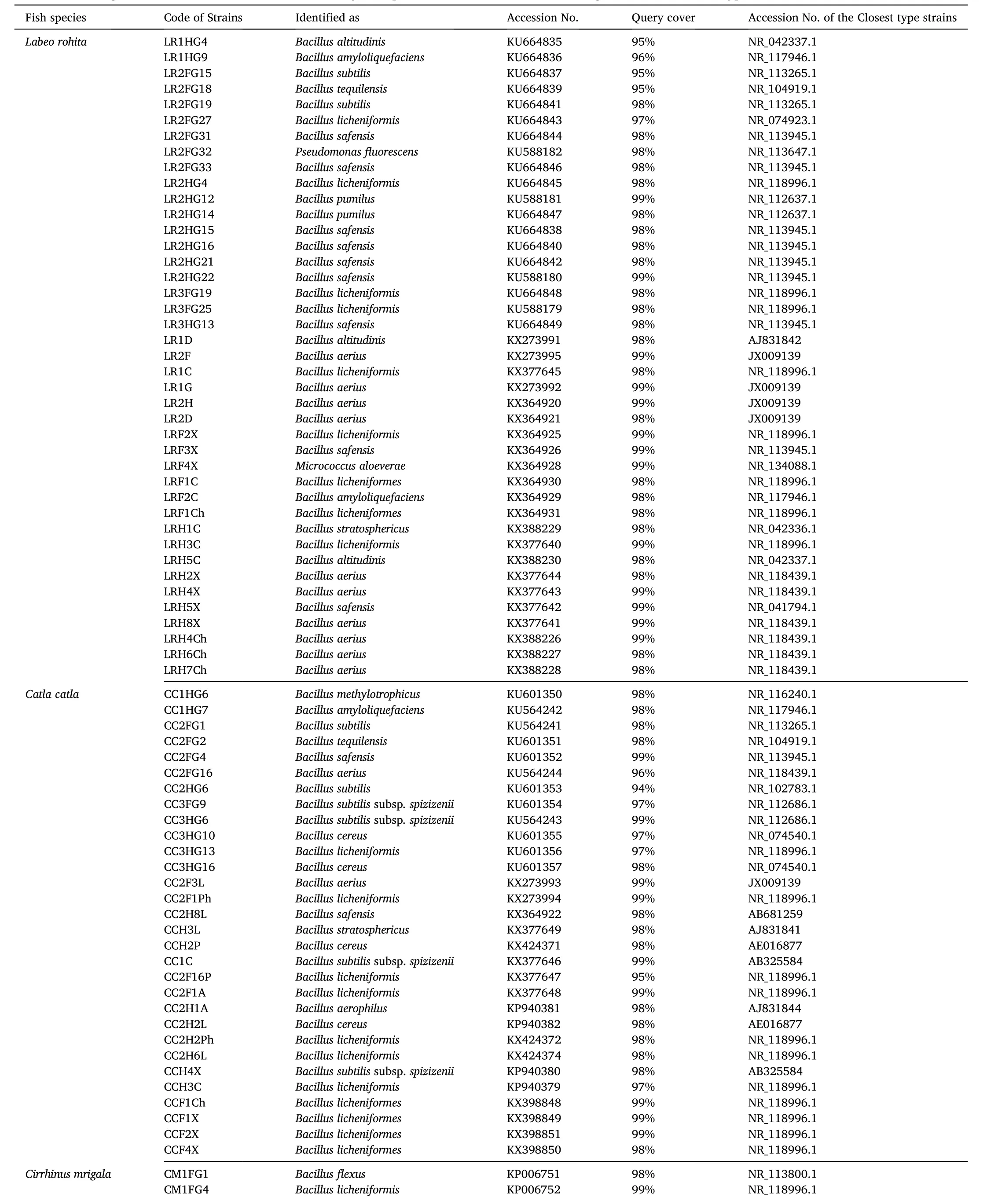
Table 2Identification of gut bacteria isolated from three Indian Major Carps, viz., L. rohita, C. catla and C. mrigala with their closest type strains retrieved from NCBI GenBank.
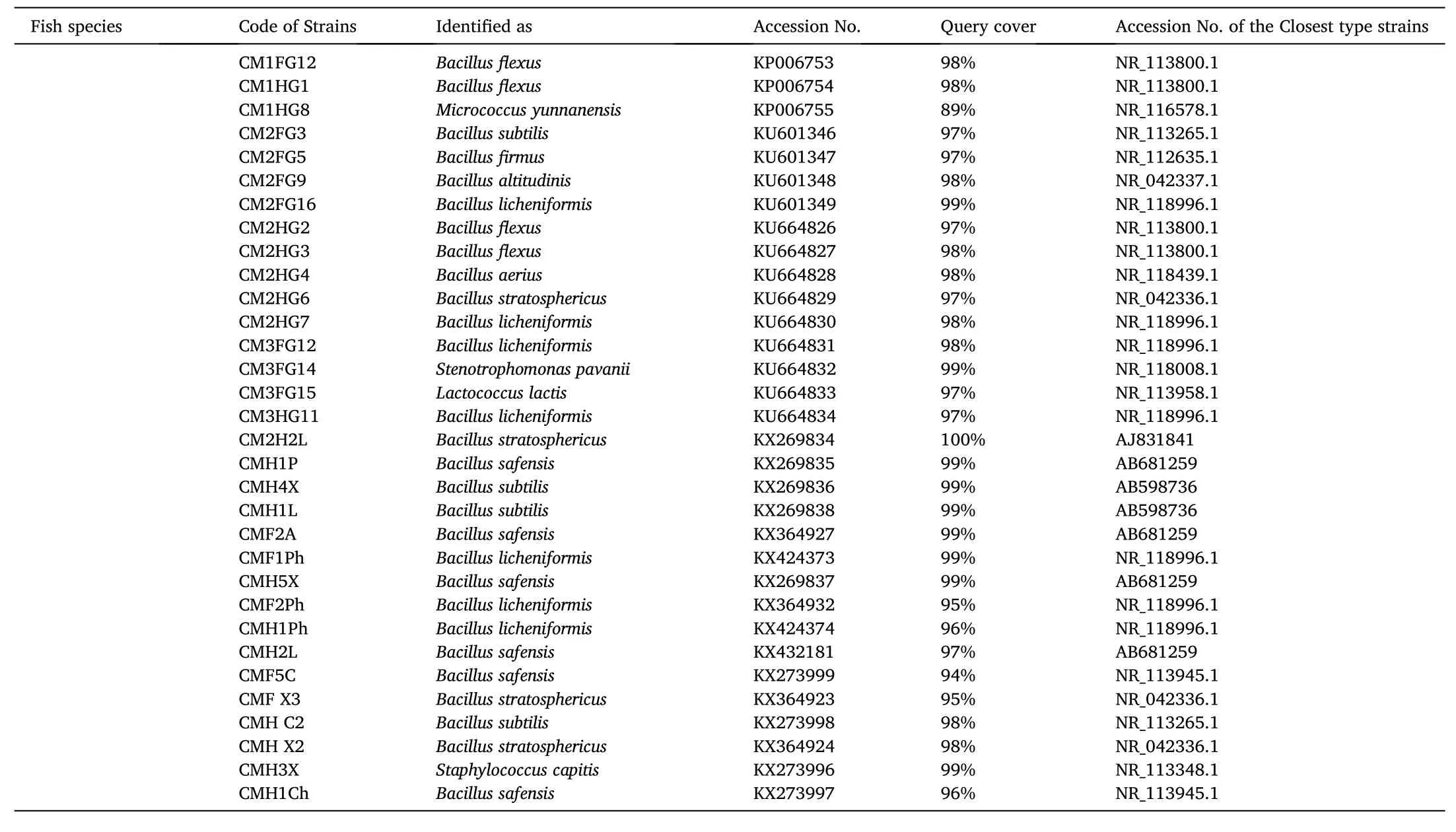
Table 2 (continued)
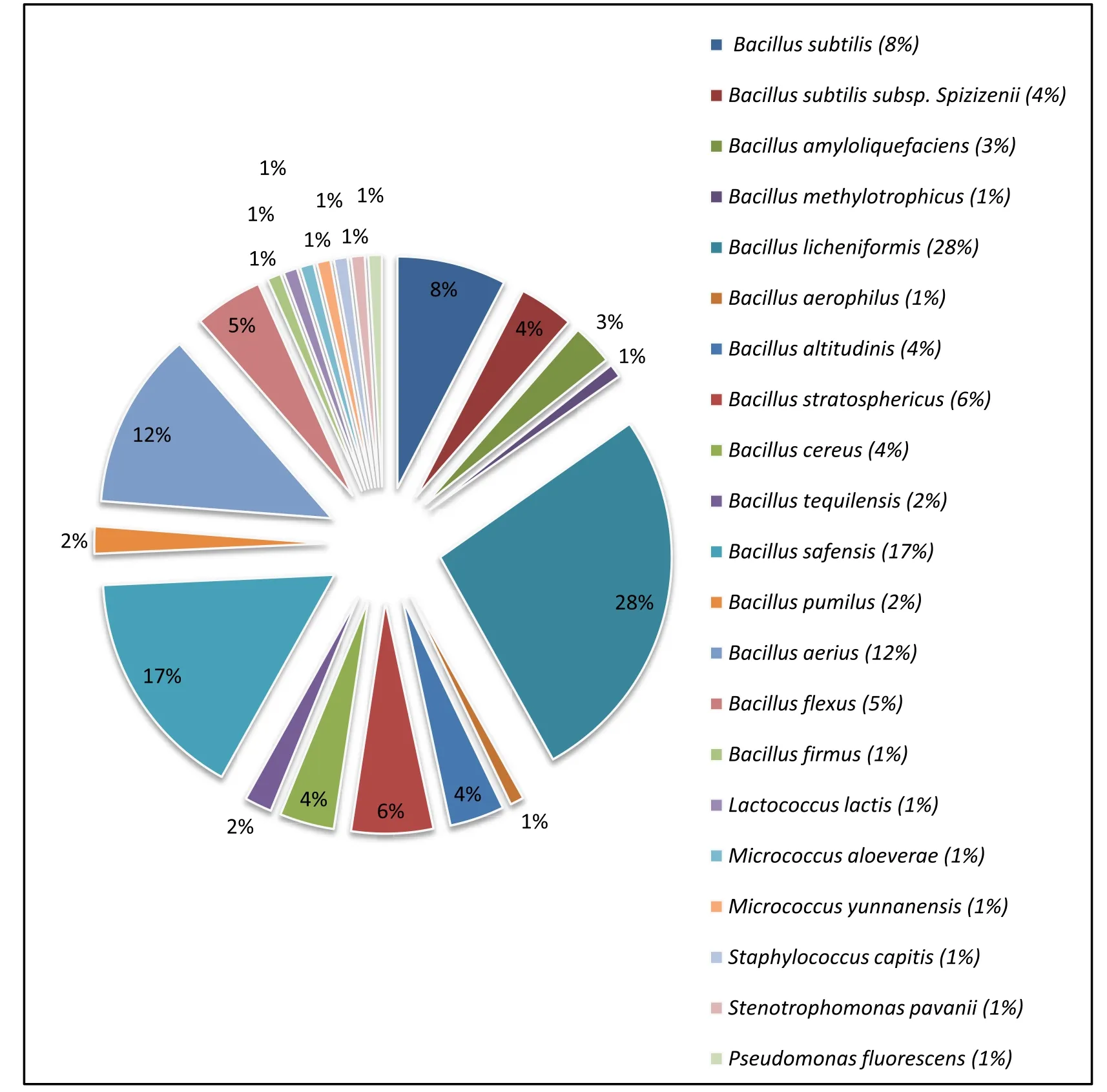
Fig. 2.Pathogen-inhibitory endosymbiotic bacteria detected in the gut of the three Indian major carps. Out of the 105 pathogen-inhibitory gut isolates, 99 strains(94.29%) belonged to the genus Bacillus.
3.3.Enzymatic activity
Results of the quantitative determination of exo-enzyme producing ability with respect to both, digestive (amylase, protease and lipase) and degradation (cellulase, phytase and xylanase) enzymes, revealedsignificant differences in the enzyme activities among the 27 primarily selected bacteria (Table 3). Amylase, protease, lipase and cellulase were produced by all of the studied bacteria, although at varying levels.Fifteen bacteria produced all six enzymes studied. Bacillus subtilis CMHC2 exhibited maximum amylase activity (271.39 ±2.14 U).Maximum protease (80.08 ± 4.15 U) and lipase (5.68 ± 0.27 U) activities were recorded with Bacillus pumilus LR2HG12 and Bacillus safensis CMF5C, respectively. Maximum cellulase (69.55 ±2.58 U) and phytase(265.42 ±5.22 U) activities were noticed with the strain Bacillus subtilis subsp. spizizenii CC1C. Phytase-producing ability by the strains B. licheniformis LR2FG27, Pseudomonas fluorescens LR2FG32,B. licheniformis LR2HG4, B. licheniformis LR3FG25, B. aerius LR2F,B. stratosphericus CCH3L, B. cereus CCH2P, Stenotrophomonas pavanii CM3FG14, B. safensis CMH1P and B. subtilis CMH4X were not recorded.Bacillus aerius LRH8X demonstrated the highest xylanase activity (13.28±1.27 U), while xylanase activity was not detected with the strains B. altitudinis LR1D, B. licheniformis LR3FG19 and B. licheniformis LR3FG25.
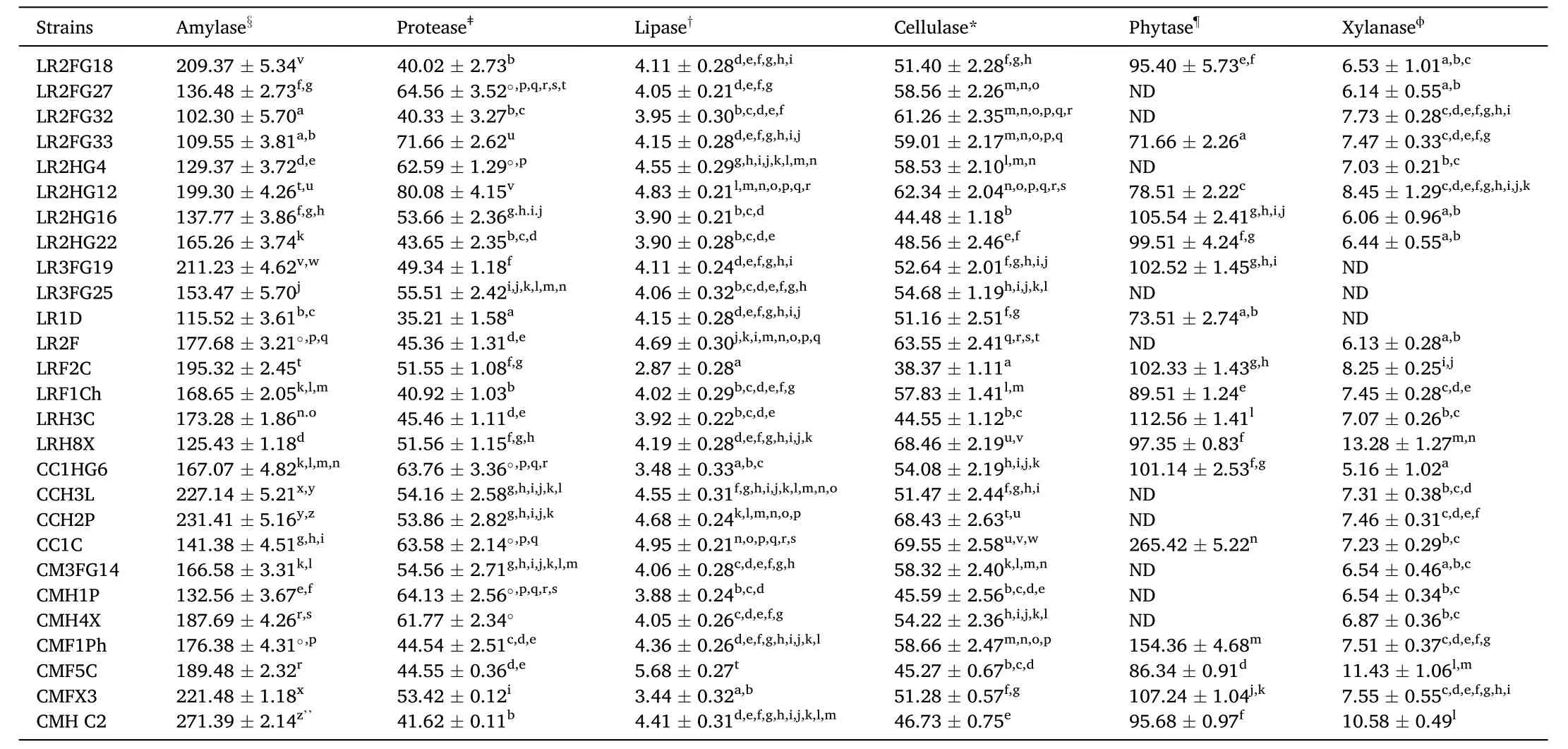
Table 3Spectrum of extracellular enzyme production by the selected bacteria. Data are means ±SE (n =3). Values with the same superscripts in the same vertical column are not significantly different (P <0.05).
3.4.Stability in gut micro-environment
3.4.1.Growth in mucus
Log viable cell counts (Log CFU/mL) revealed that the 27 primarily selected strains were competent to grow in both, intestinal mucus as well as skin mucus (Table 4). In general, the strains were more potent to grow in intestinal mucus than the skin mucus. The lowest growth (6.38 ±0.01 Log CFU/mL) was revealed by B. licheniformis LRF1Ch in skin mucus,while the highest growth (7.32 ±0.10 Log CFU/mL) was recorded for B. licheniformis LR2HG4 grown in mucus collected from intestine.
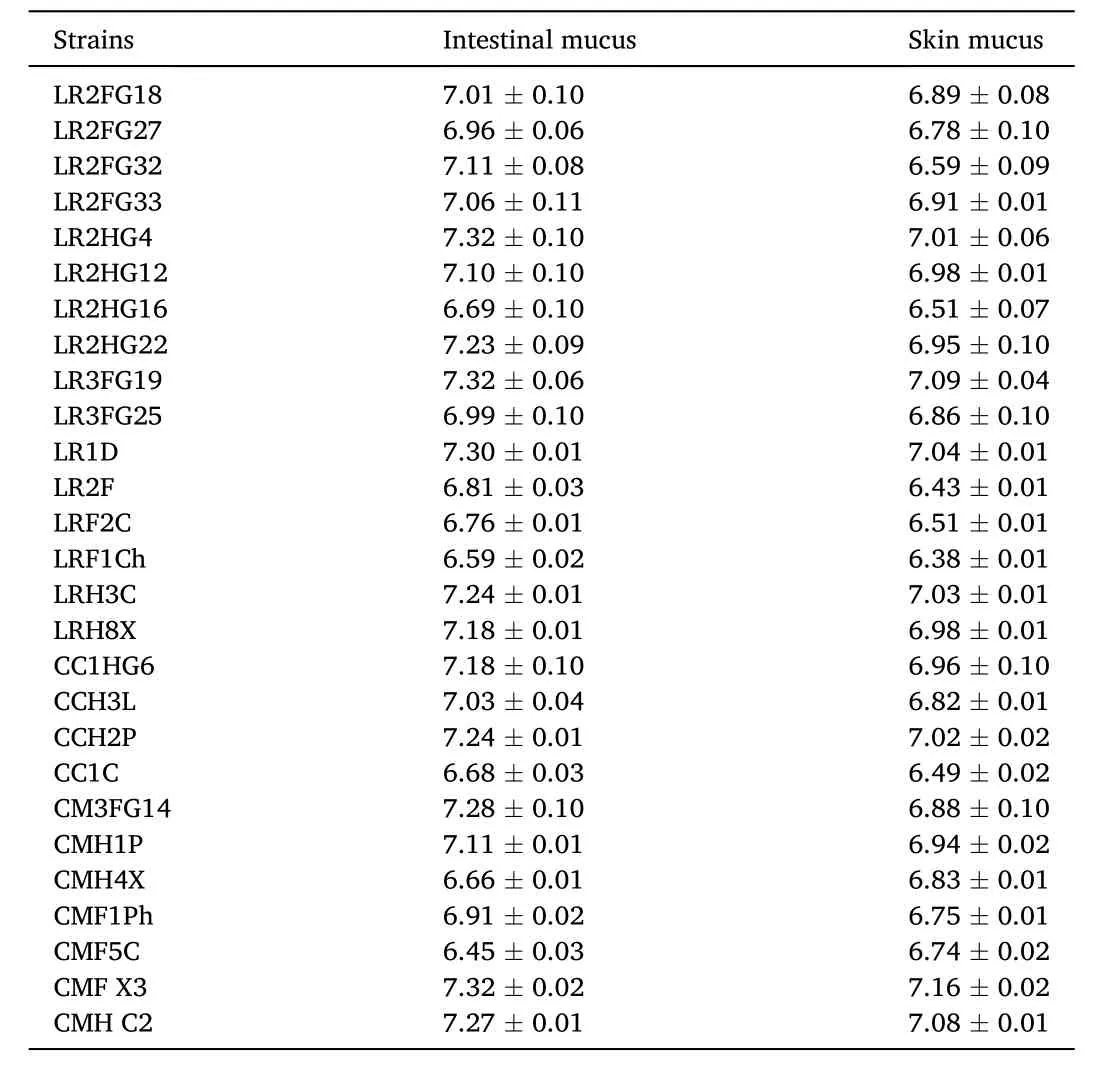
Table 4Log values of viable count (Log CFU mL-1) of the selected gut bacteria (Initial count: 6 Log CFU mL-1 mucus) grown in skin and intestinal mucus of carps.Viable count was done on TSA plates inoculated with respective bacteria cultures of 24h in fish mucus. Data are mean ±SE (n =3). No growth detected on plates inoculated with sterilized mucus.
3.4.2.Tolerance to bile juice
All of the primarily selected strains exhibited tolerance towards diluted (20%) bile juice (pH 5.5-7), even after exposure for a period of 24h. Growth detected on TSA plates inoculated with bacteria suspension treated with 20% bile has been presented in Table 5. The highest and lowest growth potential in terms of viable counts (Log CFU/mL) have been recorded for the strains Bacillus pumilus LR2HG12 (7.23 ±0.01)and Bacillus subtilis subsp. spizizenii CC1C (6.12 ±0.01), respectively.

Table 5Tolerance of the selected gut bacteria at different concentrations of fish bile juice for 1.5 h at 30 °C. Viable count was determined on TSA plates inoculated with bile exposed bacterial suspension. Data are mean ±SE (n =3).
3.5.Bio-safety assays
3.5.1.Hemolytic assay
Hemolytic activities of the primarily selected strains are shown in Table 6. When grown on blood agar media plates, 11 strains produced greenish halo under or around the colonies and thus were categorized as α-hemolytic. The strains B. licheniformis CMF1Ph and P. fluorescens LR2FG32 were considered as β-hemolytic as they produced clear halo around colonies. Another 14 strains, B. tequilensis LR2FG18,B. licheniformis LR2HG4, B. pumilus L R2HG12, B. safensis LR2HG22,B. altitudinis LR1D, B. licheniformis LRH3C, B. aerius LRH8X,B. methylotrophicus CC1HG6, B. stratosphericus CCH3L, Bacillus cereus CCH2P, S. pavanii CM3FG14, B. safensis CMH1P, B. stratosphericus CMFX3, and B. subtilis CMHC2 did not produce halo zone around the colonies indicating that these strains were γ-hemolytic. Only bacteria with γ-hemolytic property were selected for the antibiotic susceptibility assay.
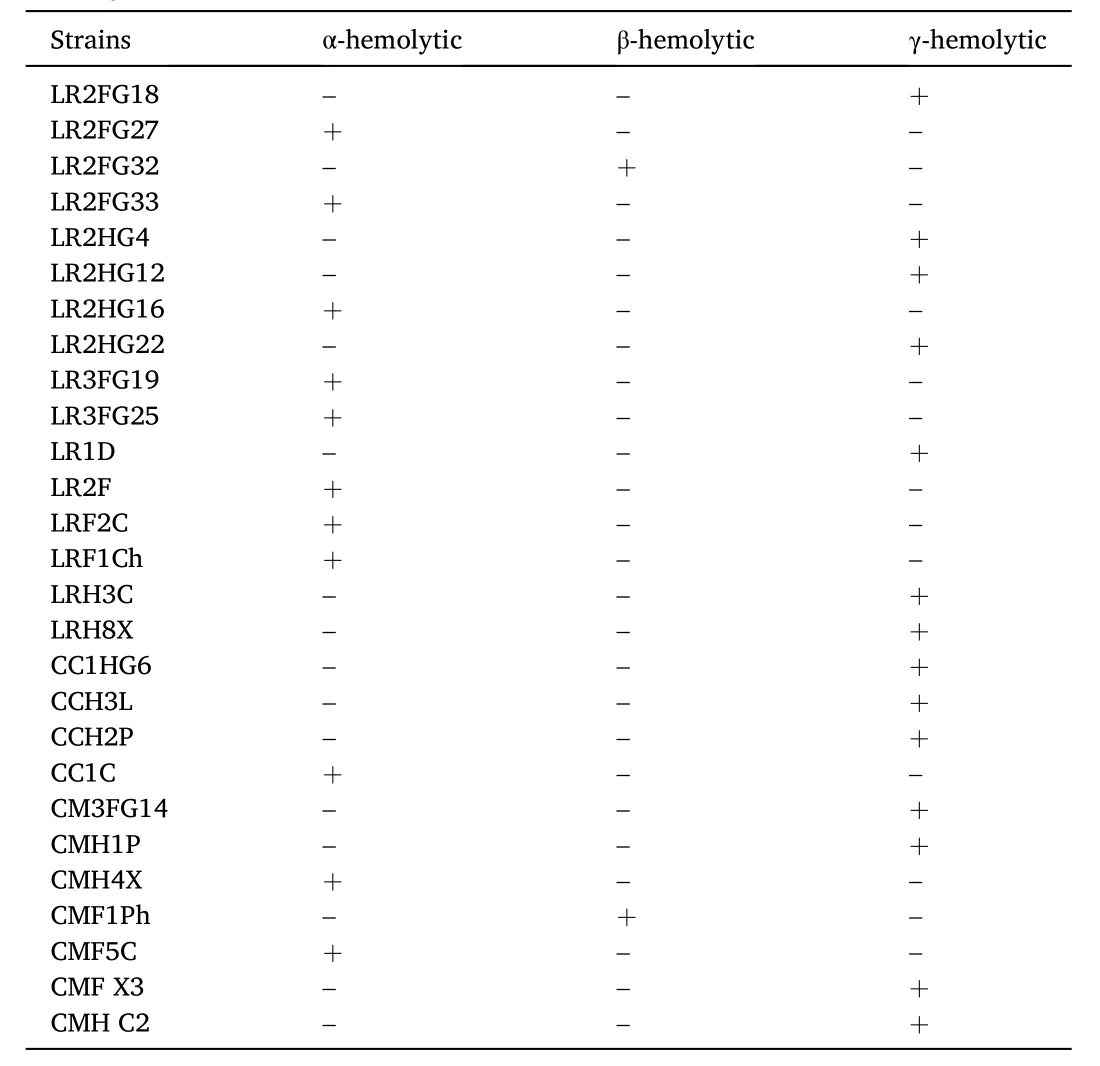
Table 6Hemolysis assay of the selected bacteria performed on Columbia blood agar base supplemented with goat blood. Appearance of hemolytic zones were classified as: α (greenish halo), β (clear halo) or γ (no halo).
3.5.2.Determination of antibiotic susceptibility
The finally selected 14 γ-hemolytic bacteria were evaluated for susceptibility against 14 antibiotics, and were noticed as susceptible to 13 commonly used antibiotics (Table 7). All of the studied bacteria were only intermediately susceptible to the amoxycillin (Am) (10mcg), while no resistance was revealed against the other tested antibiotics.
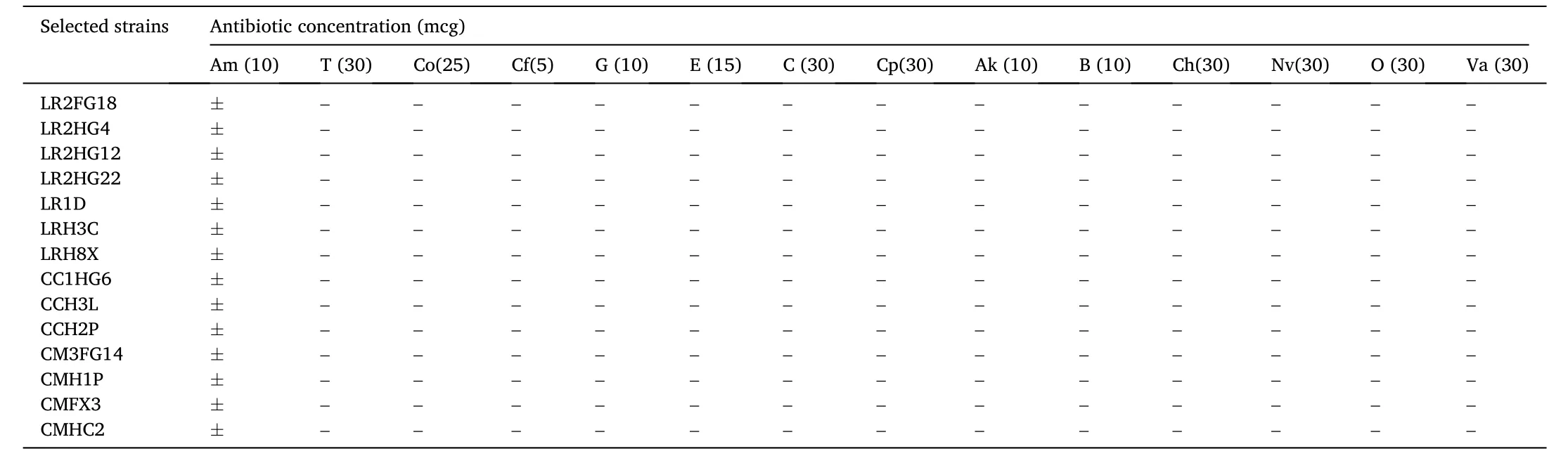
Table 7Antibiotic sensitivity of the selected bacteria.
3.5.3.Small-scale in vivo validation
At the end of the 4 weeks trial following intra-peritoneal injection, no external anomalies, disease symptoms or mortality was revealed in the control group, as well as in the experimental groups (results not shown).Consequently, the selected bacteria appeared as harmless to the fingerlings of Indian major carps.
4.Discussion
The presently reported study demonstrated diversity of the autochthonous pathogen-inhibitory gut bacteria in three IMCs reared under polyculture system and portrayed their likely probiotic potential. The study used culture based-techniques for isolation of autochthonous gut microbiota, however, universal primers guided amplification and analyses of the 16S rRNA partial gene sequences were employed to identify the pathogen-inhibitory gut microbiota. Commonly, the use of conventional culture-based techniques is argued because of lacking accurateness, requiring more time, and being incapable to represent a correct picture of the bacterial diversity as majority of the microorganisms are unculturable (Asfie, Yoshijima, & Sugita, 2003; Egerton, Culloty,Whooley, Stanton, & Ross, 2018; Gajardo et al., 2016; Li et al., 2015;Ray, Roy, Mondal, & Ring?, 2010). Thus, culture dependent methods based on 16S rRNA gene sequences using universal primers may not reproduce the core diversity of a given environment (Gajardo et al.,2016; Marchesi et al., 1998; Suzuki & Giovannoni, 1996), including the gut microenvironment. On the contrary, it may be apprehended that the presence of any bacterium would not suggest its functional role (Ray,Ghosh, & Ring?, 2012), e.g. antagonistic or enzymatic potential, within the gut. Therefore, as the major aim of the present study was to decipher pathogen-inhibitory gut bacteria in the IMCs, the use of a culture-based technique seemed to be logical.
The presently reported study revealed that pathogen-inhibitory bacterial community in the three IMCs were almost similar being dominated by Bacillus spp., which were in accordance with previous reports on gut bacterial community in freshwater teleosts (Ghosh et al.,2010; Ray et al., 2010; Mondal, Roy, & Ray, 2010). Occurrence of Bacillus spp. in the GI tract of finfish and shell fish, and their probiotic potential in aquaculture has been widely investigated (for review, see Soltani et al., 2019; Kuebutornye, Abarike, & Lu, 2019). Although,pathogen inhibition by gut bacteria in fish has been less studied, likely antagonism against different fish pathogens has been suggested to be considered as one of the desired criteria in the probiotic screening process during recent times (Dutta, Banerjee, Mukherjee, & Ghosh,2018; Mohapatra, Chakraborty, Kumar, de Boeck, & Mohanta, 2013;Mukherjee et al., 2017, 2019b; Nandi et al., 2018). In accordance to the present study, B. subtilis SG4 isolated from C. mrigala showed antagonistic activity against fish pathogenic P. fluorescens, A. hydrophila and E. tarda (Ghosh, Sinha, & Sahu, 2007). Pathogen inhibition by bacilli isolated from gastrointestinal (GI) tract of rohu, L. rohita (Giri et al.,2011) has been reported. Probiotic B. subtilis BT23 and Bacillus spp.could inhibit growth of pathogenic Vibrio harveyi, both in vitro and in vivo (Janarthanam, George, John, & Jeyaseelan, 2012; Vaseeharan &Ramasamy, 2003). The antagonistic activity of Lactobacillus casei andLactobacillus plantarum
, isolated from common carp (Cyprinus carpio
)intestines was studied usingin vitro
double agar layer method againstYersinia ruckeri
(Andani, Tukmechi, Meshkini, & Sheikhzadeh, 2012).The presently reported study also noticed the presence of pathogen-inhibitory lactic acid bacteria (Lactococcus lactis
CM3FG15) in the gut of mrigal. However, due to low cumulative inhibition score(<
10),L. lactis
CM3FG15 was not included in further characterization of probiotic properties. Furthermore, strains ofB. aerius
,B. sonorensis
(Dutta, Banerjee, Mukherjee, & Ghosh, 2015) andB. methylotrophicus
(Mukherjee & Ghosh, 2016) isolated from catla andB. cereus
andB. circulans
isolated from the GI tract of some other fish species (Lalloo,Moonsamy, Ramchuran, G¨orgens, & Gardiner, 2010; Geraylou et al.,2014) were established as antagonistic against diverse strains of pathogenicA. hydrophila
. Strains ofB. methylotrophicus
isolated from channel cat fish (Ictalurus punctatus
) intestine inhibited fish pathogens causing enteric septicaemia (Edwardsiella ictaluri
) and motile aeromonad septicaemia (Aeromonas hydrophila
) (Ran et al., 2012). In another study,B. subtilis
isolated from the GI tract of channel cat fish inhibitin vitro
growth ofA. hydrophila
,A. sobria
, andA. caviae
,in vitro
(Luo, Bai, &Chen, 2014); while a strain ofB. sonorensis
CM2H3L isolated from the gut ofC. mrigala
was reported to inhibit pathogenicA. salmonicida
(Dutta & Ghosh, 2015). Mukherjee et al. (2016) documented strains ofB. stratosphericus
,B. aerophilus
,B. licheniformis
andSolibacillus silvestris
isolated from the GI tract of mrigal inhibitedin vitro
growth of someAeromonas, Psudomonas
andBacillus
fish pathogens. Inhibition of pathogenic aeromonads by diverse strains of bacilli (B. methylotrophicus
,B. amyloliquefaciens
,B. licheniformis
) isolated from rohu were also revealed by Mukherjee et al. (2017) and Dutta et al. (2018) (B. subtilis
subsp.spizizenii
,B. tequilensis
).Although the present study detected antagonism of gut-associated bacteria against some fish pathogens, the mechanism of inhibition was not addressed. Preceding reports indicated that inhibitory activity of bacteria could be due to single or combined production of anti-microbial compounds, e.g., antibiotics (Williams & Vickers, 1986), bacteriocins(Desriac et al., 2010; Pisano et al., 2014), siderophores, lysozymes,proteases and hydrogen peroxide (Sugita, Fujie, Sagesaka, & Itoi, 2009).The double agar layer method used in the present study to evaluate the antagonistic effect of the putative probiotics detects the in fluence of diffusing antimicrobial substances on the growth of pathogenic bacteria(Kesarcodi-Watson et al., 2008). Thus, the present study confirmed the production of one or more antibacterial substances by fish gut bacteria isolates inhibitingin vitro
growth of fish pathogens. In the present investigation, 105 out of 1216 bacterial isolates depicted inhibitory activity againstA. hydrophila
,A. salmonicida
andA. sobria
, which are most common among diseases causing by bacteria of freshwater fish(Mukherjee et al., 2017). Another pathogen used in the present study,P. fluorescens
, has been described as an opportunistic pathogen of various fish species (B?gwald & Dalmo, 2014). Thus, the presently reported study might support the hypothesis that antagonism between endogenous fish gut bacteria (i.e. “normal” or protective microbiota)and pathogens constitute a major component of ‘defensive barrier function
’ in fish (Cain & Swan, 2010, pp. 111-134; P′erez-S′anchez et al.,2011; Sahoo, Jena, Patel, & Seshadri, 2016; Hoseinifar et al., 2018;Mukherjee et al., 2019b).Another beneficial property for the selection of probiotics is;enzyme-producing abilities (Dutta et al., 2015; Kesarcodi-Watson et al.,2008; Merrifield et al., 2010), as extracellular enzyme producing bacteria in fish gut exert positive effects to the digestive processes of the host (Ray et al., 2012). The present study demonstrated extracellularamylase, protease and lipase producing abilities, and anti-nutritional factors degrading-enzymes; cellulase, phytase and xylanase of selected bacterial strains. Presence of amylolytic, proteolytic, cellulolytic and lipolytic bacteria in the GI tracts of tropical freshwater fish has been widely studied (Banerjee, Dora, & Chowdhury, 2013; Mondal et al.,2010; Ray et al., 2012). Ghosh, Sen, and Ray (2002) isolated Bacillus spp.strains from in the gut of rohu which were good protease and cellulase producers. Likewise, Mondal et al. (2010) isolated amylolytic, cellulolytic and proteolytic B. licheniformis and B. subtilis from the digestive tract of bata (Labeo bata). Ray et al. (2010) detected a large population of amylase, cellulase and protease producing bacteria in the gastrointestinal (GI) tract of three Indian major carps, catla, mrigal and rohu.However, none of these studies considered the antimicrobial potential of the enzyme producers. Previously, few reports dealt with both enzymatic and pathogen-inhibitory potential of the gut bacteria in tropical freshwater fishes (Dutta et al., 2015, 2018; Kavitha, Raja, & Perumal,2018; Midhun et al., 2017; Mukherjee et al., 2017; Mukherjee & Ghosh,2016), which were in accordance with the present report. Based on the present findings, we put forward the hypothesis, that the exoenzyme-producing bacteria colonizing within the GI tract of studied freshwater fish might offer protection against some fish pathogens or vice versa.
Furthermore, the potential of the isolates to colonize the intestine was assessed in terms of their capacity to grow in fish intestinal mucus in vitro. All of the 27 primarily selected gut isolates could grow in intestinal mucus. However, minor differences were recorded in bacterial growth rate within mucus, which might be due to specific nutritional requirements of the bacteria or differences in oxygen concentration or pH of the medium as suggested by Geraylou et al. (2014). Vine, Leukes, and Kaiser (2004) determined high growing rate of five candidate probiotics bacteria isolated from the gut of the common clown fish (Amphiprion percula), while Geraylou et al. (2014) isolated different strains of Bacillus spp. and Lac. lactis having promising probiotic characteristics from the gut microbiota of Siberian sturgeon (Acipenser baerii). Among the isolates, Lactococcus lactis ssp. lactis STG45 and STG81 showed high viability and highest adhesion capacity to fish intestinal mucus. In accordance to the recent observations by Mukherjee and Ghosh (2016)and Dutta et al. (2018), the presently reported study also revealed that intestinal mucus is a good growth medium for the selected putative probiotics. Therefore, it might be apprehended that the putative probiotics characterized in the present report are likely to survive through the fish GI tract and colonize.
Bile usually exhibits specific and non-specific defense mechanisms of the gut against harmful bacteria (Charteris, Kelly, Morelli, & Collins,1998). Therefore, higher tolerance to fish bile is an important criterion for the selection of a potential aquaculture probiotic to ensure their survival and growth in the small intestine of fish (Balc′azar et al., 2008).In the present investigation, the primarily selected strains showed tolerance to high concentrations of bile (20% bile juice, pH 5.5-7). This finding may be related to the ability of the tested isolates to reduce the inhibiting effect of the bile salt via bile salt hydrolase (BSH) activity (De Smet, Hoorde, Woestyne, Christiaens, & Verstraete, 1995). Bile tolerance of probiotic bacteria intended for aquaculture application has been revealed in several studies (Buntin, Chanthachum, & Hongpattarakere,2008; Dutta et al., 2015; Mukherjee & Ghosh, 2016; Nikoskelainen et al.,2001).
Among the 27 primarily selected isolates that showed antagonism towards pathogens as well as promising exoenzymes producers were subsequently tested for haemolytic activity. In the present investigation,14 out of 27 isolates were hemolysis-negative strains (γ-hemolytic).Non-hemolytic strain of B. subtilis was reported by Nayak and Mukherjee(2011). Although there is no such correlation between haemolysis and the pathogenic nature of bacteria (Inamura, Muroga, & Nakai, 1984;Kodama, Moustofa, Mikami, & Izawa, 1985), haemolytic strains should be avoided as harmful haemolytic bacteria may cause serious infection in the skin and mucous membranes (Madigan, Martinko, & Parker,2000; Ouwehand, Salminen, & Isolauri, 2002).
Intensive aquaculture has encouraged the growth of different infectious disease of fish, which leads to a raise in the use of different antimicrobial agents and chemotherapeutics (Defoirdt, Sorgeloos, & Bossier,2011). However, fish do not actively metabolize antibiotic and pass them back to environment. According to Burridge, Weis, Cabello,Pizarro, and Bostick (2010), 75% of the antibiotics applied with feed are released into the water. Moreover, uncontrolled use of antibiotics in aquatic environment induces a selective pressure for emergence of drug-resistant bacteria, and thus, it was strictly criticized and restricted(Romero et al., 2012; S?rum, 2006). The emergence of multidrug-resistant pathogens leading to sudden infectious disease outbreaks is the most challenging problem in the aquaculture industry,resulting in heavy economic loss (Thankappan, Ramesh, Ramkumar,Natarajaseenivasan, & Anbarasu, 2015). Therefore, an ideal probiotic must be free of any plasmid-encoded antibiotic resistance genes (Merrifield et al., 2010). In the present study, antibiotic sensitivity of the selected 14 hemolysis-negative bacterial isolates was determined using 14 broad spectrum antibiotics commonly used in aquaculture. All of the strains were highly susceptible to the tested antibiotics, except for amoxycillin (10 mcg). The presence of antibiotic sensitive strains within the fish gut was in accordance with the previous studies (Nayak &Mukherjee, 2011; Thankappan et al., 2015; Sahoo et al., 2016). In contrast, Kim and Austin (2008) reported a broad spectrum of antibiotic resistance of Carnobacterium strains with probiotic properties isolated from rainbow trout (Oncorhynchus mykiss) intestine.
The selection of probiotic candidate should be given high precedence as inappropriate microorganisms might cause an imbalance in the ratio of good-to-bad bacteria (for review see, Lazado, Caipang, & Estante,2015). The presently reported study might propose a selection criterion of the probiotics from autochthonous source that covers functional characterization, gut stability and bio-safety. Previous reports have indicated that probiotics could be linked to the modulation of gut microbiota in fish (Allameh et al., 2016; G′omez & Balc′acar, 2008;Merrifield et al., 2010). Specifically, application of the probiotics might lead to alteration of microbial diversity (Ramos et al., 2013) and decrease in the abundance of pathogenic organisms (Pereira, Pereira,Soares, Mouri~no, & Merrifield, 2019) within the gut. Thus,pathogen-inhibitory gut bacteria characterized as putative probiotics in the present report might be considered as a tool for manipulation of gut micro flora in the hosts, as indicated elsewhere (Andani et al., 2012;Asaduzzaman et al., 2018). However, further studies are warned on this particular issue.
5.Conclusion
The present report pretends to help the aquaculture industry and scientific community by expanding the knowledge on potentiality and composition of the culture dependant gut bacteria in the IMCs. Among the pathogen inhibitory symbiotic bacteria detected in the present study, only 14 isolates were noticed with excellent exo-enzyme producing ability, γ-hemolytic and sensitive to the commonly used antibiotics. At present, there is an effort to emphasize host-associated microbiota as probiotics for the aquaculture industry (Lazado et al.,2015). Thus, the presently reported study might offer a scope to consider gut-associated bacteria as putative probiotics for the carps. A single probiotic candidate may not be used for all fish species, as a single strain may become ineffective for a particular species (Asaduzzaman et al.,2018; Lazado et al., 2015). These 14 isolates represented by genus Bacillus (13 nos.) and Stenotrophomonas (1 nos.), might form a probiotic consortium for prospective use in composite carp culture. However,further studies involving feeding trials with large number of fish in different commercial farm conditions are needed prior to application of these consortia in aqua-farming.
CRediT authorship contribution statement
Koushik Ghosh: Conceptualization, Supervision, Data curation,Writing - original draft, Writing - review & editing. Anjan Mukherjee:Investigation, Methodology, Formal analysis, Software. Dipanjan Dutta: Investigation, Methodology. Sudeshna Banerjee: Investigation,Methodology. Eva Marie Breines: Methodology. Ellinor Hareide:Methodology. Einar Ring?: Conceptualization, Writing - review &editing.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
Authors are grateful to the Head, Department of Zoology, The University of Burdwan, West Bengal, India; University Grants Commission(UGC-SAP-DRS programme), New Delhi, India and Department of Science and Technology (FIST and PURSE programmes), New Delhi, India for research support.
 Aquaculture and Fisheries2021年2期
Aquaculture and Fisheries2021年2期
- Aquaculture and Fisheries的其它文章
- Editorial: Global fish passage issues
- Analysis of the impacts of socioeconomic factors on hiring an external labor force in tilapia farming in Southern Togo
- Using Bayesian Bio-economic model to evaluate the management strategies of Ommastrephes bartramii in the Northwest Pacific Ocean
- Expression of multi-domain type III antifreeze proteins from the Antarctic eelpout (Lycodichths dearborni) in transgenic tobacco plants improves cold resistance
- A chromosome-level genome assembly of the red drum, Sciaenops ocellatus
- Loss of scleraxis leads to distinct reduction of mineralized intermuscular bone in zebra fish
