Effects of enrichmemt planting with native tree species on bacterial community structure and potential impact on Eucalyptus plantations in southern China
Yuxing Xu ·Chao Li·Wankuan Zhu ·Zhichao Wang ·Lichao Wu·Apeng Du
Abstract Multi-generational planting of Eucalyptus species degrades soil quality but the introduction of legumes can improve soil fertility and microbial diversity.However,the effects ofintroducing non-legume native tree species on soil nutrients and bacterial community structure remain poorly understood.This study investigated the impacts of the conversion of third generation monoculture Eucalyptus plantations to mixed systems including Eucalyptus urograndis with Cinnamomum camphora (EC) and E.urograndis with Castanopsis hystrix (EH), on soil chemical and biochemical properties and bacterial community structure, diversity and functions.First generation E.urophylla plantations were the control.Results show that planting the third generation Eucalyptus led to a significant decrease in pH, organic matter, nutrient content, enzyme activities (invertin, acid phosphataes, and urease), and bacterial α-diversity compare to the controls.However, the mixed planting showed significant improvement in soil chemical and biochemical attributes and bacterial α-diversity, although the E.urograndis and C.hystrix planting had no improvement.Chloroflexi (oligotrophic bacteria) were significantly enriched in third generation Eucalyptus and Eucalyptus + C.hystrix, while proteobacteria increased significantly in the E.urograndis with C.camphora plantings.The relative abundance of multiple metabolic pathways increased significantly in the third generation Eucalyptus plantations whereas membrane transportrelated genes were enriched in soils of the mixed systems.The changes in bacterial community structures in the two mixed systems were driven by diversity, organic matter and acid phosphatase, while bacterial functions were affected by invertase, NO-3-N, diversity and urease.These results suggest that the transformation of successive monoculture Eucalyptus plantations into mixed plantations reduces the depletion of soil nutrients and enhances the ecological function of soil microorganisms.
Keywords Soil chemical propertie·Enzyme activity ·16S rRNA amplicon sequencing·Bacterial community structure·Tax4Fun function prediction
Introduction
Eucalyptusspecies are widely planted in southern China,and their wood is an important industrial raw material (Gon?alves et al.2013).In China, as of 2019, the area ofEucalyptusplantations exceeded 5.4 × 106ha, and continues to increase (Arnold et al.2020).However, large areas were established as monocultures, with multi-generation continuous planting, which has caused numerous ecological and environmental problems, e.g., soil degradation and loss of ecosystem stability (Liu and Li 2010; Xu et al.2020).These problems pose a serious threat to the sustainable management of plantations (Arnold et al.2020) and prompt rethinking what sustainableEucalyptusmanagement is.Is it truly sustainable as it is?
Inappropriate management measures may be one of the main driving forces leading to the decline of soil quality(Tu et al.2017b; Pereira et al.2019; Zhu et al.2021).Introducing native tree species into monoculture plantations is generally considered to be an effective measure to address this problem (Piotto et al.2004; Chu et al.2019).Previous initiatives have focused on a mixture ofEucalyptusand legume species (e.g.,Acacia mangium) (Koutika et al.2017,2019).However, based on the demand for different types of material in the wood processing industry, ever more large state-owned forest farms are gradually introducing non leguminous native species intoEucalyptusplantations (Guo et al.2018).Many non-legumes, broad-leaved species, such asCinnamomum camphora(L.) J.Presl.andCastanopsis hystrixMiq., have been introduced into monocultureEucalyptusplantations due to their excellent wood properties and adaptability to humid and acidic soil (Guo et al.2018; Xu et al.2021).
Research on mixed plantations is increasing and the mixed-mode approach indicates high levels of ecological function relative to monocultures (Amazonas et al.2018).Koutika et al.( 2017) investigated the soil of monocultureEucalyptusplantations,Acaciaplantations, and mixed plantations and found that the mixedEucalyptus×Acaciaplantations significantly increased the turnover rate of soil nitrogen (N) and phosphorus (P) and promoted microbial activity.The mixed plantation as a good allocation model,can not only improve soil nutrient status and enzyme activities, but also significantly increase the proportion and stability of soil aggregates, which have a good buffering capacity to cope with extreme environmental changes (Laclau et al.2008; Zagatto et al.2019).However, there are relatively few studies on the mixed effects ofEucalyptusand non-legume native species, including in-depth examination of related mechanisms of soil improvement.
Microorganisms respond to environmental changes through altered community structure, and microbial diversity and functional changes are indicators that reflect changes in land use and soil fertility status (Steenwerth et al.2003; Tu et al.2017a; Zheng et al.2020).As the main components of soil microbial communities (Fierer and Jackson 2006),bacteria communities are closely related to the dynamic changes of nitrogen (N), starting from the process of N fixation and extending to N mineralization and soil greenhouse gas emissions (Nelson et al.2016).In addition, soil bacterial communities are closely related to the soil phosphorous(P) cycle.Studies have confirmed that some soil bacteria possess the ability to mineralize inorganic and organic P by secreting organic acids and phosphorus hydrolase (Maougal et al.2014; Wan et al.2020).Therefore, it is necessary to evaluate the composition, structure, and function of the soil bacterial communities in different plantations (monoculture or mixed).Rachid et al.( 2013) and Epron et al.( 2016)and noted that, due to the symbiotic N fixation relationship between leguminous plants and rhizobia, the mixed planting ofEucalyptusand leguminous species form a unique underground bacterial community structure which facilitates N and P cycling.However, there is relatively little research on how a mixture ofEucalyptusand non-legume tree species affect soil bacterial communities, thereby affecting nutrient cycling, absorption and losses.
In this study, soil bacterial community structure and functions were evaluated in first and third monoculture generations ofEucalyptusand two intercropped systems (E.urograndiswithC.camphoraandE.urograndiswithC.hystrix), which resulted from a third rotation of aEucalyptusplantation.It was hypothesized that: (1) multi-generation continuous planting ofEucalyptusleads to a decline in soil nutrients and enzyme activities, and results in changes in the structure and function of the bacterial communities; (2)mixed plantation management improves soil nutrient status,which is reflected in the diversity of bacteria and the relative enrichment of some functional bacteria; and, (3) changes in soil biochemical indicators caused by the transformation measures will drive changes in soil microbial community structure and function.
Materials and methods
Study area
The study area was located at the Guangdong ZhanjiangEucalyptusPlantation Ecosystem Research Station, China(21°16′ N, 110°05′ E, Fig.S1).This site has a maritime monsoon climate with an average temperature of 23.1 °C,and a long- term average annual rainfall of 1385 mm,concentrated from May to September.The area is flat and belongs to the terraced terrain of the platform and low hills.
The soil type is Rhodi-Udic Ferralosols according to the Chinese Soil Taxonomy Classification (Gong 1999), which derives from shallow sea sediments.In 1990, the Leizhou Forestry Bureau began to plantAcacia mangiumWilld.in this area.Ten years later felling of this plantation was carried out.In 2002, the China Eucalypt Research Centre (CERC)established a first-generationEucalyptus urophyllaplantation; most of the plantation was harvested June 2010.A second generation of the hybridEucalyptus urophylla×E.grandiswas established in August 2010 and harvested December 2015.After digging planting holes and applying a base fertilizer, the experimental design started in June 2016.
Experiment design and soil sampling
Approximately 10 ha of selected cutover land were divided into four equal blocks with an isolation zone between each block (July 2016).Three plantings were randomly arranged in each block.The first planting (E) was continuous planting of the third generation monocultureEucalyptus urophylla×E.grandis.The second planting (EC) was a mixed plantation ofE.urograndisandC.camphoraat the same planting density as E.The third planting (EH) was a mixed plantation ofE.urograndisandC.hystrixat the same planting density.Four adjacent first-generationE.urophyllaplantations (without management) were selected as controls(CK).Eucalyptusseedlings were 6-months old, 15 - 20 cm in height, and provided by the South China Experiment Nursery.Seedlings of native tree species were one year old,100 - 150 cm in height, and cultivated in the nursery of the local forestry science institute.Before reforestation, mechanical digging (50 cm × 50 cm × 40 cm) was used at 3-m and 2-m spacing.A 0.25 kg of special base fertilizer (N: P2O5:K2O=7: 12: 6; HY-6–12-6, Huayang Environmental Protection Technology Co., Ltd., Zhaoqing, China) was applied per hole, for a total of 416.7 kg per ha.Each treatment was augmented with a compound fertilizer (N: P2O5: K2O=15:6: 9; HY-15–5-6) as topdressing after one year.Topdressing was within 1-m of the trunk at 0.5 kg per tree, for a total of 833 kg per ha.
Fifteen soil samples in the 10-cm layer were randomly taken from each replicate of these four plantings using a soil extractor and thoroughly mixed after removing the humus and debris (December 2019).A total of 16 mixed soil samples were obtained from the four plantings.From each of the 16 samples, approximately 2-g fresh soil (sieved through 2-mm mesh) was placed in a 2 mL centrifuge tube and steeped into liquid N for subsequent amplicon sequencing.On the day of collection, the frozen samples were transferred to refrigerator storage at - 80 °C until needed for analysis.A remaining 1-kg was divided into two equal parts, one part transferred to a refrigerator at 4 °C for enzyme activity analyses, and the other air-dried for a week and used for soil chemical analysis.
Soil chemical properties and enzyme activity analysis
The pH of each sample was determined with an electronic pH meter (soil: water 1:2.5; Reijonen et al.2016).Soil organic matter (OM) was determined by the potassium dichromate-sulfate colorimetric method (Sims and Haby 1971).Total nitrogen was measured with the Kjeldahl method (Tsiknia et al.2014), and NH+4-N and NO-3-N determined by 2 mol·L-1KCL leaching-indophenol blue colorimetric method and UV spectrophotometry, respectively (Lu 1999).Total phosphorus was determined by the sodium hydroxide fusion-molybdenum antimony colorimetric method (Liu et al.2016) and available phosphorus measured by the hydrochloric acid-ammonium fluoride extraction-molybdenum antimony colorimetric method (Lu 1999).
Soil urease activity (URE) was determined by measuring the amount of ammonium released from the soil (Kandeler and Gerber 1988); acid phosphatase (ACP) and invertin(INV) were measured by the phenylphosphonium-4-aminoantipyrine colorimetric method and the 3,5-dinitrosalicylic acid colorimetric method, respectively (Lu 1999).
Soil bacterial DNA extraction and sequencing
Bacterial community genomic DNA was extracted from soil samples using the E.Z.N.A.? soil DNA Kit (Omega Bio-tek,Norcross, GA, U.S.) according to manufacturer’s instructions.The DNA extract was checked on 1% agarose gel, and DNA concentration and purity determined with NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA).To improve the accuracy of species classification, this study selected the highly recognized V3–V4 regions of the 16S rRNA gene as the amplification region with primer pairs 338F (5’- ACT CCT ACG GGA GGC AGC AG-3’) and 806R (5’-GGA CTA CHVGGG TWT CTAAT-3’)(Mori et al.2013).PCR amplification of the 16S rRNA gene was performed using ABI GeneAmp? 9700 and the product extracted from 2% agarose gel and purified using the Axy-Prep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to manufacturer’s instructions and quantified using Quantus? Fluorometer.The NEXTFLEX Rapid DNA-Seq Kit for Illumina (BIOO) was used to generate sequencing libraries and then applied to an Illumina Miseq PE300 platform for sequencing (Illumina, San Diego,CA, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co.Ltd.(Shanghai, China).
Processing of sequencing data
Trimmomatic and FLASH were used to quality-filter and merge the raw fastq files of 16 woodland soil samples.Sequences exhibiting ≥ 97% similarity were assigned to the identical operational taxonomic unit (OTU) by UPARSE software (version 7.1, http:// drive5.com/ uparse/) (Edgar 2013).To minimize the effects of sequencing depth on subsequent community composition analyses, the number of sequences from each sample was rarefied to the smallest size.The taxonomy of each OTU representative sequence was aligned against the 16S rRNA database ( https:// www.arb- silva.de/) with a confidence threshold of 0.7 according to the RDP Classifier algorithm (11.1 http:// rdp.cme.msu.edu/).After the classification, the self-written Perl script was adopted to count phyla, classes, orders, families, and genera.Three indices of the alpha diversity, including Chao1, Shannon index and Simpson index, were calculated using Mothur software (1.30.2, https:// www.mothur.org/ wiki/ Downl oad_mothur).
Statistical analysis
Principal coordinates analysis (PCoA) of Bray–Curtis distance was performed to compare the community structure dissimilarities among samples using the ‘a(chǎn)pe’ and ‘vegan’packages with R version 3.4.0 (R Core Team 2019).To evaluate the effects of plantation composition on bacterial community structure, a permutational multivariate analysis of variance (PERMANOVA) was carried out using the‘a(chǎn)donis’ function in the package ‘vegan’ in R.The linear discriminate analysis (LDA) effect size (LEfSe) method ( http://hutte nhower.sph.harva rd.edu/ galaxy/) (Segata et al.2011)was used to identify distinguishing taxa to illustrate significant differences between land use patterns in this study.An LDA threshold score > 4.0 and a significancep-value < 0.05 were set.The default value (LDA=2.0) is a widely accepted standard for Lefse analysis.However, a higher value setting can reduce the number of differences in taxa and improve the accuracy of analysis (Fisher 1936), which proves that it is reasonable to use LDA > 4 in bacterial classification analysis.
To better understand the potential functional profiles,the program Tax4Fun ( http:// tax4f un.gobics.de/) (A?hauer et al.2015) was run in R software ( https:// www.r- proje ct.org/) based on the 16S rRNA data.The SILVA-labeled OTU table was used to compute metabolic capabilities using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto 2000).Based on this information,the pathway information was obtained and the abundance of each functional category calculated according to the abundance of OTUs.
A redundancy analysis (RDA) and spearman correlation analysis were performed using Canoco 4.5 (Biometris,Wageningen, Netherlands) and SPSS (version 17.0, IBM,Armonk, NY, USA) to observe relationships between soil chemical properties, and bacterial community structure and function, respectively.Soil chemical properties, enzyme activity, α-diversity of bacterial communities, and relative abundance of genes associated with different functional categories were examined by one-way analysis of variance(ANOVA).Tukey’s HSD post-hoc tests were used to assess any significance differences among means (p< 0.05).All one-way statistical analyses were performed with SPSS (version 17.0, IBM, Armonk, NY, USA).
Results
Soil chemistry properties
Different plantation management resulted in significant differences in soil chemical properties.The continuous planting of third-generationEucalyptus(E) and the mixed plantations ofE.nurograndiswithC.hystrix(EH) significantly reduced soil OM, total N, NO-3-N, and available P compared with the first-generationEucalyptusplantations (CK) (Table 1,p< 0.05).However, mixed plantations ofE.urograndiswithC.camphora(EC) improved pH, OM, total N, total P, and available P concentrations significantly compared with E and EH plantings (p< 0.05).
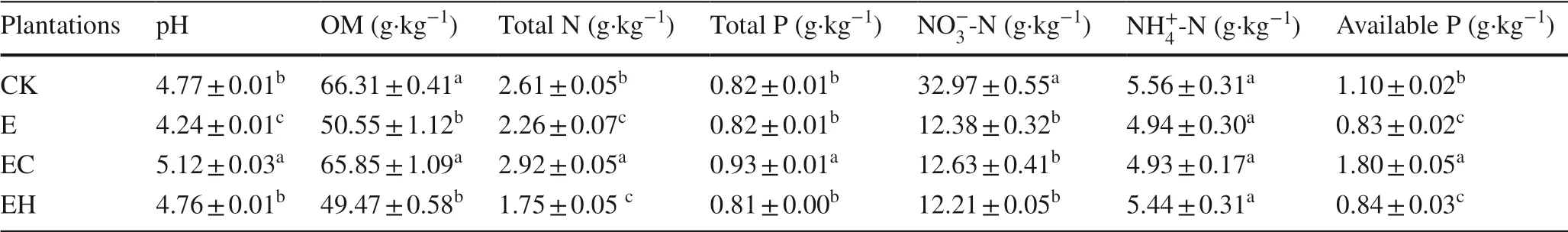
Table 1 Soil chemical properties of first rotation Eucalyptus plantations (CK), third rotation Eucalyptus plantations (E), E.urograndis with Cinnamomum camphora plantations (EC) and E.urograndis with Castanopsis hystrix plantations (EH) in Guangdong Zhanjiang Eucalyptus Plantation Ecosystem Research Station, China
Soil enzyme activity
Soil enzyme activity showed significant differences among the four plantings (p< 0.05).Specifically, changes in soil enzyme activity were closely related to changes in nutrients.INV, ACP, and URE activities in the controls (4.27 mg g-1, 0.62 mg g-1, 0.28 mg·g-1, respectively) and mixed plantations ofE.urograndiswithC.camphora(EC) (4.51 mg·g-1, 0.53 mg·g-1, 0.25 mg g-1,respectively) were significantly higher than in the thirdgenerationEucalyptusplantations (E) (1.82 mg g-1,0.39 mg·g-1, 0.24 mg·g-1, respectively) and the mixed plantations ofE.nurograndiswithC.hystrix(EH)(3.09 mg g-1, 0.40 mg g-1, 0.23 mg g-1, respectively)(Fig.1 A–C).There were no significant differences between the latter two plantings (except INV).In addition, soil enzyme activity and OM, total N, and available P concentrations showed significant positive correlations(p< 0.05, Table 2).
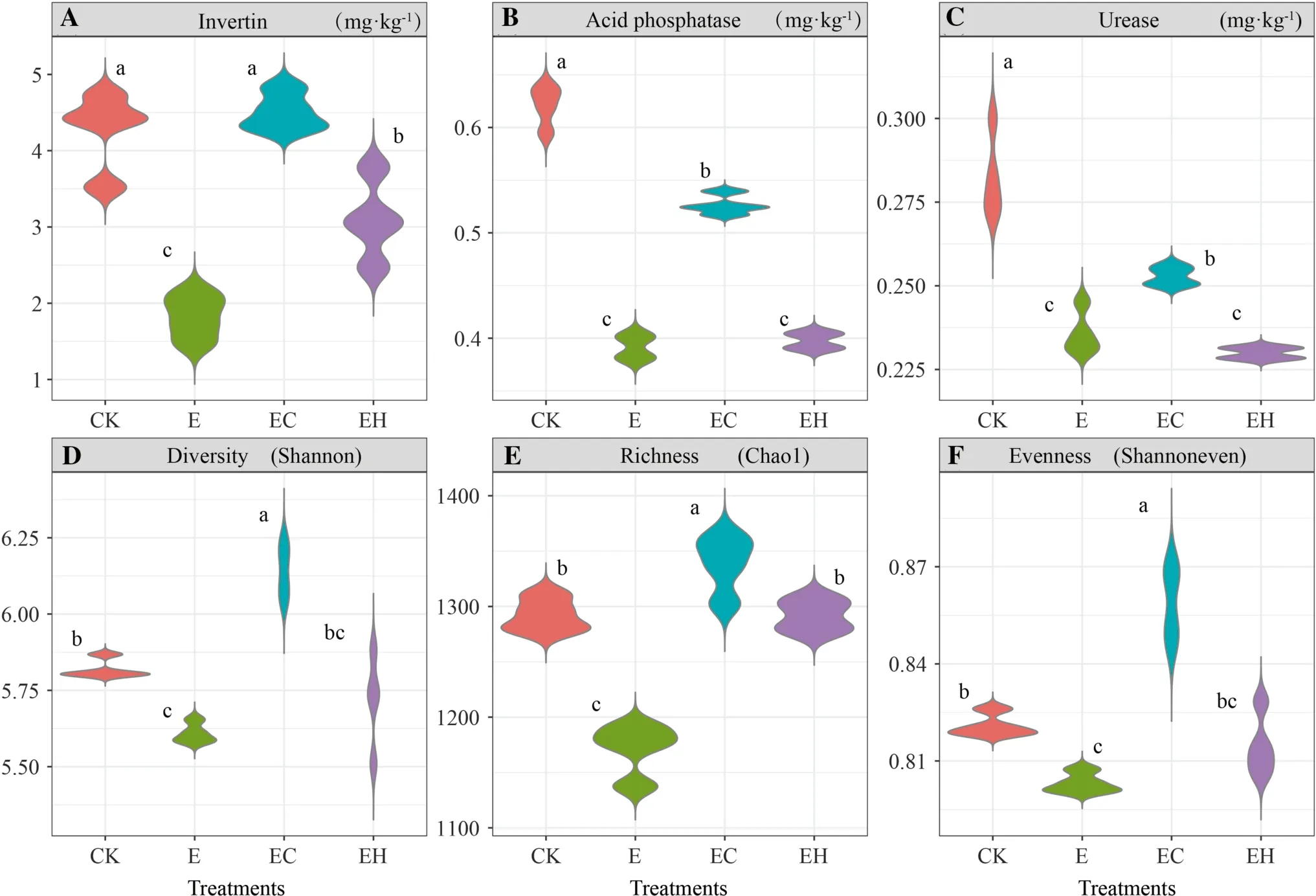
Fig.1 Soil bacterial enzyme activity and bacterial α-diversity of the first rotation Eucalyptus plantations (CK), the third rotation Eucalyptus plantations (E), E.urograndis with Cinnamomum camphora plantations (EC) and E.urograndis with Castanopsis hystrix plantations (EH) in Guangdong Zhanjiang Eucalyptus Plantation Ecosystem Research Station, China
Illumina MiSeq sequencing results and bacterial community α-diversity
A total of 760,101 quality sequences were obtained from 16 soil samples, with an average sequence length of 412 bp(Table S1).A total of 1716 OTUs were identified in all samples, of which 1195 OTUs were shared among all treatments.The lowest number of OTUs appeared in planting E (N=1359), and the highest in EC (N=1619) (Fig.S2).The soil bacteria dilution curve of the Sobs constructed by random sampling of DNA sequences tended to be flat, indicating the rationality of the sequencing data (Fig.S3).
Different plantation types caused significant changes in bacterial α-diversity (p< 0.05), which was strongly driven by soil chemical properties.The continuous planting of the third-generationEucalyptus(E) significantly reduced soil α-diversity indexes (Chao index, Shannon diversity index,Shannon evenness index).The establishment of the mixed plantations (EC, EH) improved soil microbial diversity,richness, and evenness, in which the EC planting reached a significant level (p< 0.05, Figs.1 D, E, and F).Spearman correlation analysis showed that the Chao index, Shannon index, and Shannon evenness index were positively correlated with pH, total N, and available P (p< 0.05), but not with total P,, andconcentrations (p> 0.05,Table 2).
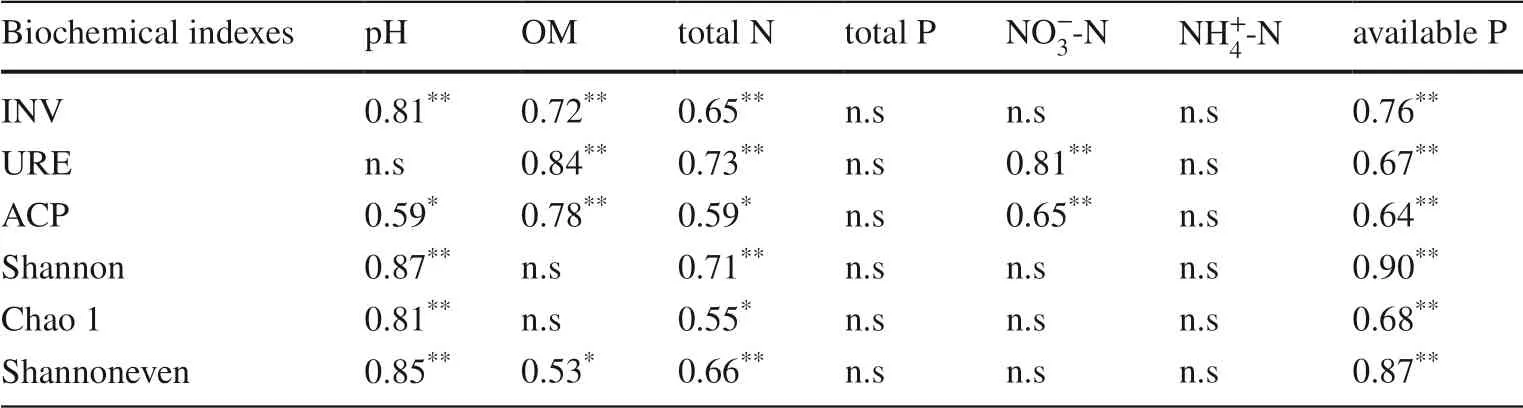
Table 2 Correlation between soil chemical properties and bacterial diversity and enzyme activity of soil samples in Guangdong Zhanjiang Eucalyptus Plantation Ecosystem Research Station,China
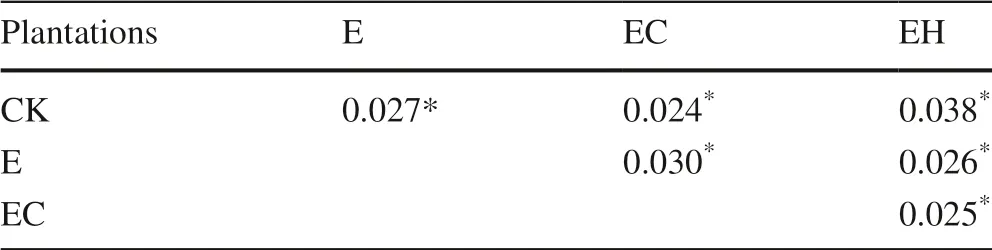
Table 3 Pairwise comparison of the soil bacterial community structure between each treatment in the PERMANOVA analysis.Data are p values
Microbial community structure and composition
The PCoA showed that there were obvious differences in the soil bacterial community structure in the different plantations, and pairwise comparison using PERMANOVA confirmed significant differences in community composition between the CK and E, EH, and EC plantings (p< 0.05, Fig.2, Table 3).Although the soil bacterial communities of E and EH were clustered, there were still significant differences (Table 3,F= 5.24,p= 0.026).The change inEucalyptusspecies, combined with the continuous planting, led to differences in soil bacterial communities between the first generationE.urophylla(CK), the third generationE.urograndis(E), and the other mixed plantations (EC and EH) (Fig.2).
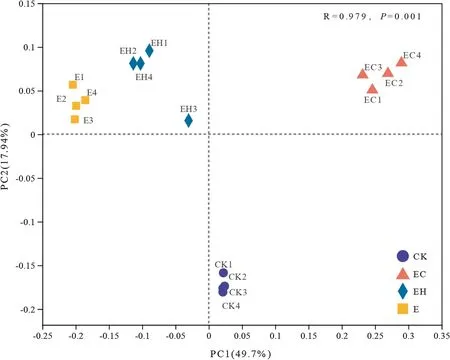
Fig.2 Principal coordinate analysis (PCoA) based on the distance matrix calculated using the weighted unifrac algorithm for 16 soil samples of the first rotation Eucalyptus plantations (CK), the third rotation Eucalyptus plantations (E),E.urograndis with C.camphora plantations (EC) and E.urograndis with C.hystrix plantations (EH) in Guangdong Zhanjiang Eucalyptus Plantation Ecosystem Research Station, China
Changes in the relative abundance of soil bacteria at each classification level reflected the impact of different plantations on the structure of bacterial communities.At the phylum level, the top three abundances were Chloroflexi, Acidobacteria and Proteobacteria (Fig.3 A).The abundance of Chloroflexi was E > EH > CK > EC.The abundance of Acidobacteria and Proteobacteria was EC > CK > EH > E.The following dominant phyla were Actinobacteria, Verrucomicrobia, WPS-2, Bacteroidetes,Gemmatimonadetes, GAL15, and Planctomycetes.In addition to the top 10 phyla with a relative abundance > 1%, the remaining 11 phyla are in “others” (Fig.3 A).
Thirteen species of class-level bacteria with relative abundance of more than 1% in different plantings were distributed in eight phyla (Fig.3 B).The most abundant classes were Acidobacteria, AD3, and Actinobacteria, which accounted for 16.5–18.4%, 7.5–20.6%, and 13.7–15.8%, respectively.The following classes in abundance were members of Proteobacteria (Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria),Chioroflexia (Ktedonobacteria and TK10), Acidobacteria(Subgroup_6), Bacteroidetes (Bacteroidia), and Verrucomicrobia (Verrucomicrobiae) (Fig.3 B).
The most abundant order was norank_c_AD3 (Chloroflexi), which accounted for 14.5%, 20.6%, 7.5%, and 20.3%in CK, E, EC, and EH, respectively, followed by three members of Acidobacteria, i.e., Acidobacteriales, Frankiales and Solibacterales, which represented 8.0–9.0%, 5.4–7.6%,and 5.1–6.9% in all samples.Moreover, the relative abundance of Rhizobiales (Proteobacteria) in four plantings was:EC > CK > EH > E.Ktedonobacterales and norank_c_TK10(Chloroflexi) showed an opposite trend (Fig.3 C).
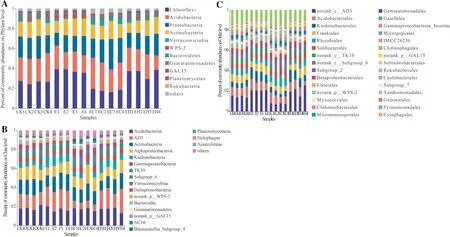
Fig.3 Soil bacterial community composition of the first rotation Eucalyptus plantations (CK), the third rotation Eucalyptus plantations (E), E.urograndis with C.camphora plantations (EC), and E. urograndis with C.hystrix plantations (EH) in Guangdong Zhanjiang Eucalyptus Plantation Ecosystem Research Station, China.A–Bacterial phylum level, B–Bacterial class level, C–Bacterial order level
For a more detailed analysis of soil bacteria, a hierarchically-clustered heat map was constructed to present species composition information and hierarchical relationships among the 50 genera shared in the 16 plantation soil samples(Fig.S4).The result of genus-level clustering was in line with the PCoA analysis, and there was a relatively close relationship between E and EH samples, significantly different from that of EC and CK.According to the heat map,most of the genus-level bacteria were similarly distributed in all soils.
Comparison of soil biomarker species in different plantations
LEfSe analysis showed that there were multiple biomarker species at different classification levels of soil bacteria in different plantings.Twenty-six bacterial clades with a significant difference were detected with an LDA threshold of 4.0 (Fig.4,p< 0.05).Bacteria were mainly subordinate to phyla Chloroflexi, Proteobacteria, Acidobacteria,and WPS-2.Specifically, Chloroflexi and WPS-2 were enriched in E, mainly containing AD3, Ktedonobacterales, TK10, and norank_p_WPS-2, while Proteobacteria were enriched in EC, mainly including Class Gammaproteobacteria, Subgroup_6, and Deltaproteobacteria.Details of the bacterial clades at the level of order, family, and genera enriched in each planting are shown in Fig.5.
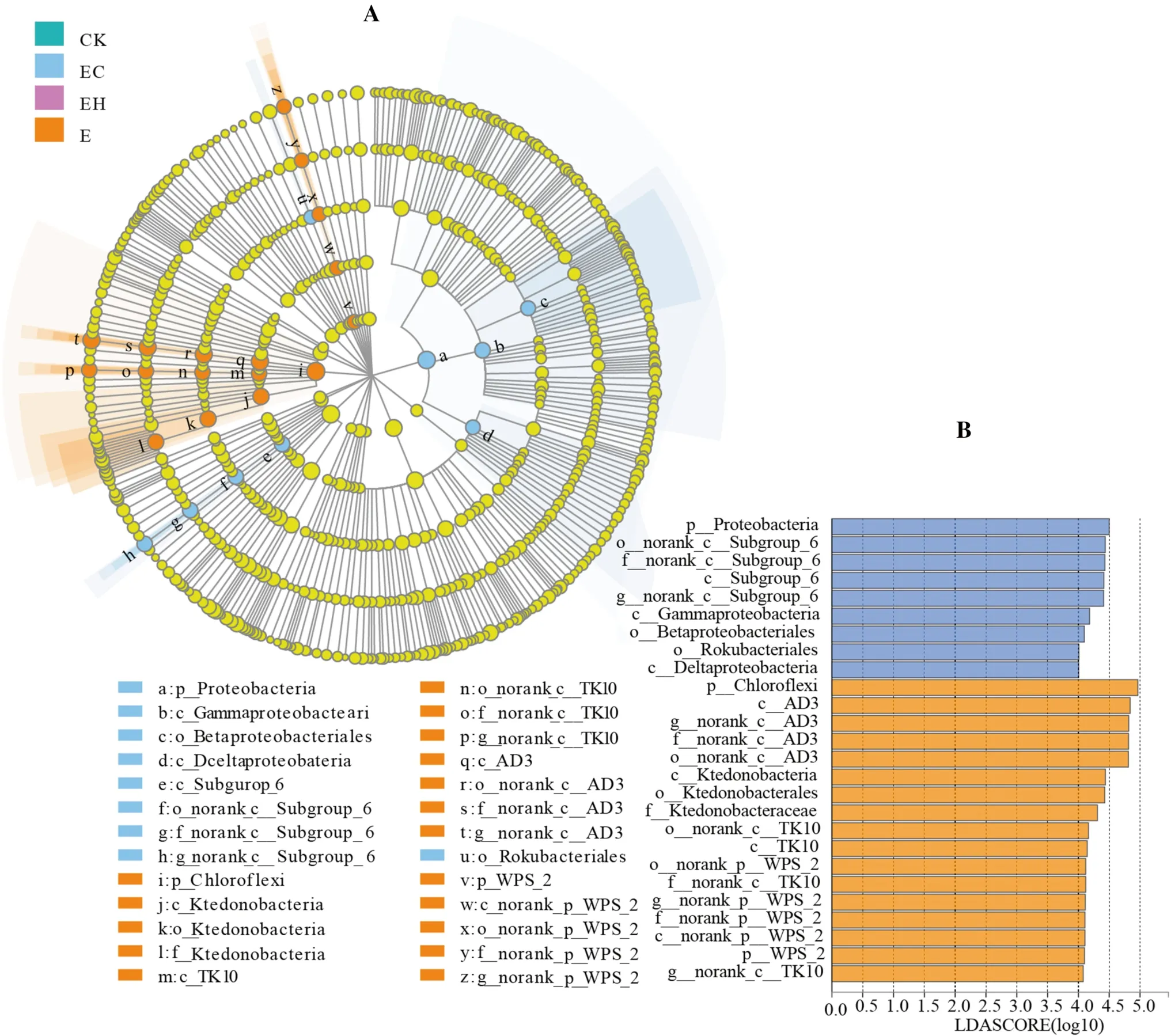
Fig.4 LEfSe analysis of soil bacterial relative abundance of the first rotation Eucalyptus plantations (CK), the third rotation Eucalyptus plantations (E), E.urograndis with C.camphora plantations (EC),and E.urograndis with C.hystrix plantations (EH) in Guangdong Zhanjiang Eucalyptus Plantation Ecosystem Research Station, China.A–Cladogram of microbial communities, B–LDA score identified the size of differentiation among different plantations with a threshold value of 4.0
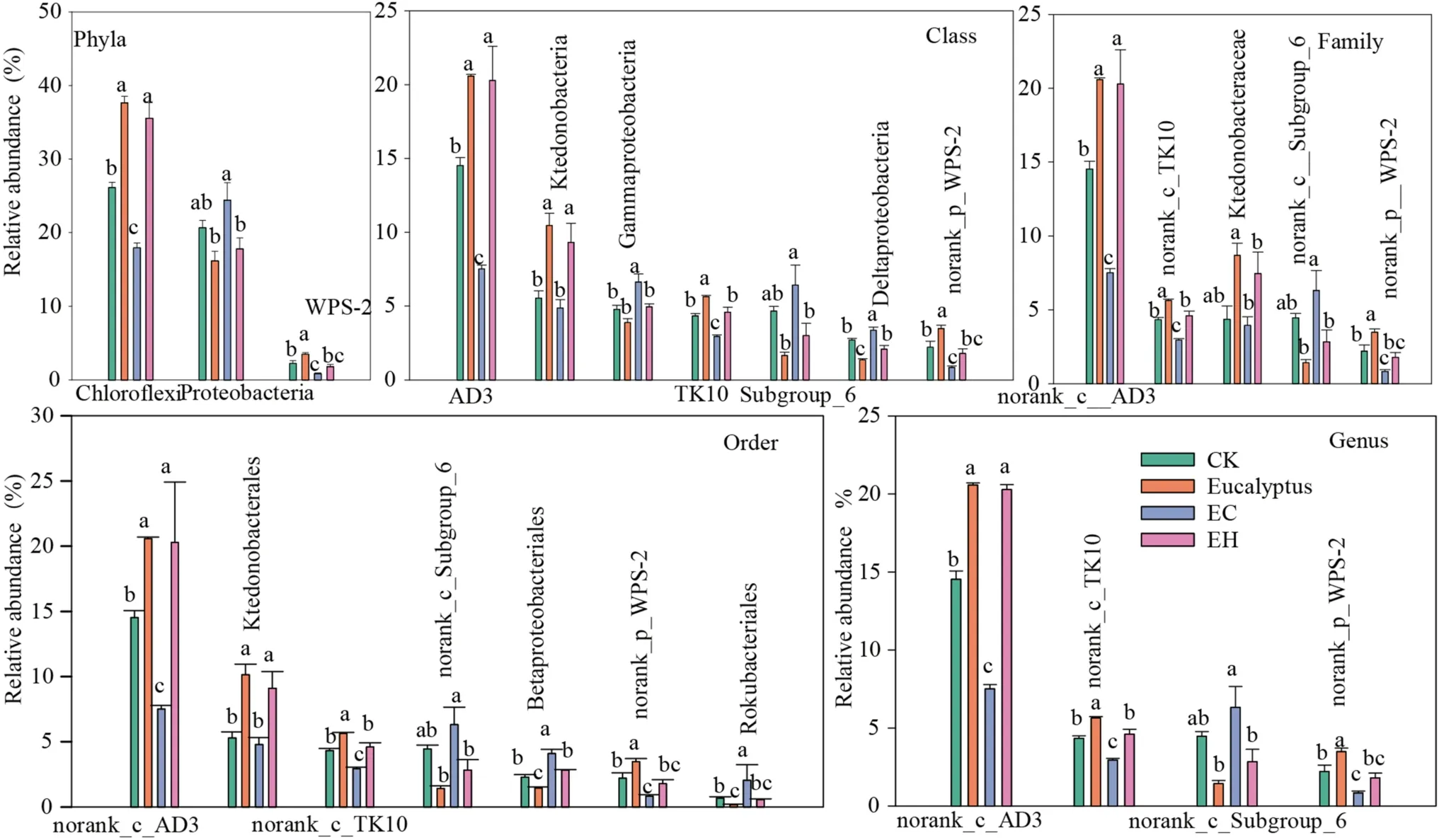
Fig.5 Comparisons of bacterial relative abundance with significant differences from phylum to genus level detected by LEfSe analysis.CK- first rotation Eucalyptus plantation soil samples, E–third rota-tion Eucalyptus plantation soil samples, EC– E.urograndis with C.camphora soil samples, EH– E.urograndis with C.hystrix soil samples
Functions of the soil bacterial community
The functional contributions of the soil bacteria in different plantation types were predicted based on 16S rRNA information using the Tax4Fun tool.The results revealed that prokaryotic microbial communities were involved in 277 predicted pathways at the functional level 3.The most abundant functional groups were ABC transporters (8.6–9.1%),two-component system (6.08–7.0%), purine metabolism(3.03–3.06%), aminoacyl-tRNA biosynthesis (2.46–2.54%),pyrimidine metabolism (1.94–2.05%), and N metabolism(1.96–2.0%) (Table S2).A total of 6191 KEGG Orthologues were obtained belonging to metabolism, environmental information processing, genetic information processing,cellular processes, and human diseases (level 1 pathways,details not listed).Of all the functional level 2 pathways, carbohydrate metabolism, amino acid metabolism, membrane transport, signal transduction, energy metabolism, metabolism of cofactors and vitamins, xenobiotics biodegradation and metabolism, nucleotide metabolism, translation, and lipid metabolism were the dominant functions (Fig.6).Interestingly, the successive planting ofEucalyptus(E) resulted in significant enrichment of multiple metabolic pathways in the soil, while the mixed plantations improved the abundance of membrane transport (especially EC, reaching a level ofp< 0.05, Fig.6).
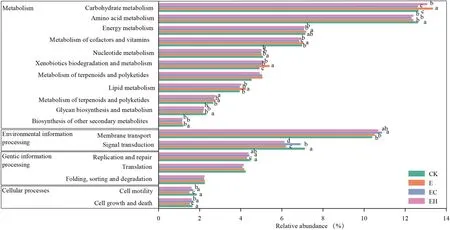
Fig.6 Variation of bacterial function profiles in soil samples for the four plantation types analyzed by Tax4Fun (function categories level 2).CK–first rotation Eucalyptus plantations, E–third rotation Euca-lyptus plantations, EC– E.urograndis with C.camphora, EH– E.urograndis with C.hystrix
Correlation between bacterial communities, functions and biochemical attributes
Soil OM, bacterial diversity, and ACP showed significant correlation with the bacterial community structure (Fig.7).These indexes showed a positive correlation with EC.Among which,soil bacterial diversity explained 66.7% of the total variation of bacterial communities (F= 27.88,p= 0.002), followed by soil organic matter (F= 4.71,p= 0.006) and ACP activity (F= 3.68,p= 0.003) (Fig.7 A).Diversity, OM, and ACP were positively correlated with Acidobacteria, Proteobacteria,Rokubacteria, and Nitrospirae, and negatively to Chloroflexi,WPS_2, and GAL15 (Table S3).
One soil chemical and three biological attributes revealed significant correlation with the bacterial potential functions profiles.Two biological attributes were positively correlated with EC, including INV (F=19.86,p= 0.002) and diversity(F= 4.23,p= 0.016), while the other two biochemical attributes showed a positive correlation with CK, including NO-3-N(F= 14.41,p= 0.002) and URE (F= 5.16,p= 0.006) (p< 0.05,Fig.7 B).Soil INV, URE, and Diversity were negatively correlated with metabolic pathways, i.e., carbohydrate metabolism,energy metabolism, metabolism of cofactors and vitamins,xenobiotics biodegradation and metabolism, metabolism of other amino acids, and lipid metabolism, but a significant positive correlation with signal transduction, replication and repair,cell motility, cell growth and cell death (Table S4).
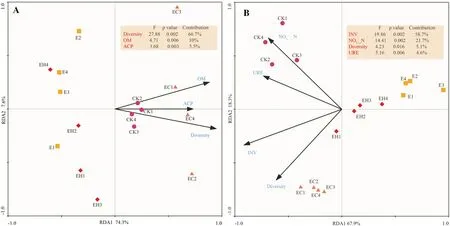
Fig.7 Redundancy analysis (RDA) based on soil biochemical attributes and dominant bacterial phylum-level relative abundance A and function profiles B of the four different plantation types (function cat-egories level 2).CK–first rotation Eucalyptus plantations, E–third rotation Eucalyptus plantations, EC– E.urograndis with C.camphora, EH– E.urograndis with C.hystrix
Discussion
Variations in soil chemical properties and enzyme activity
Soil chemical properties and enzyme activities were significantly different among the four types of plantations,and soil enzyme activities were positively correlated with soil organic matter, total N, and available P levels.After two successive generations ofEucalyptus, pH, OM, total N, NO-3-N, and available P decreased significantly in the third generation plantations (E) (Fig.8).Similar conclusions were reported in previous studies concerning soil N and P concentrations (Zhu et al.2019; Xu et al.2021).This study also found that transforming the third-generationEucalyptusplantation into a mixed system EC produced a significant improvement in soil nutrient levels compared to the mixed system EH and the third-generationEucalyptusplantations(E), while EH and E nutrient levels were insignificant.The results may be interpreted as a consequence of the higher nutrient demand of the EH and the monoculture plantations than EC.
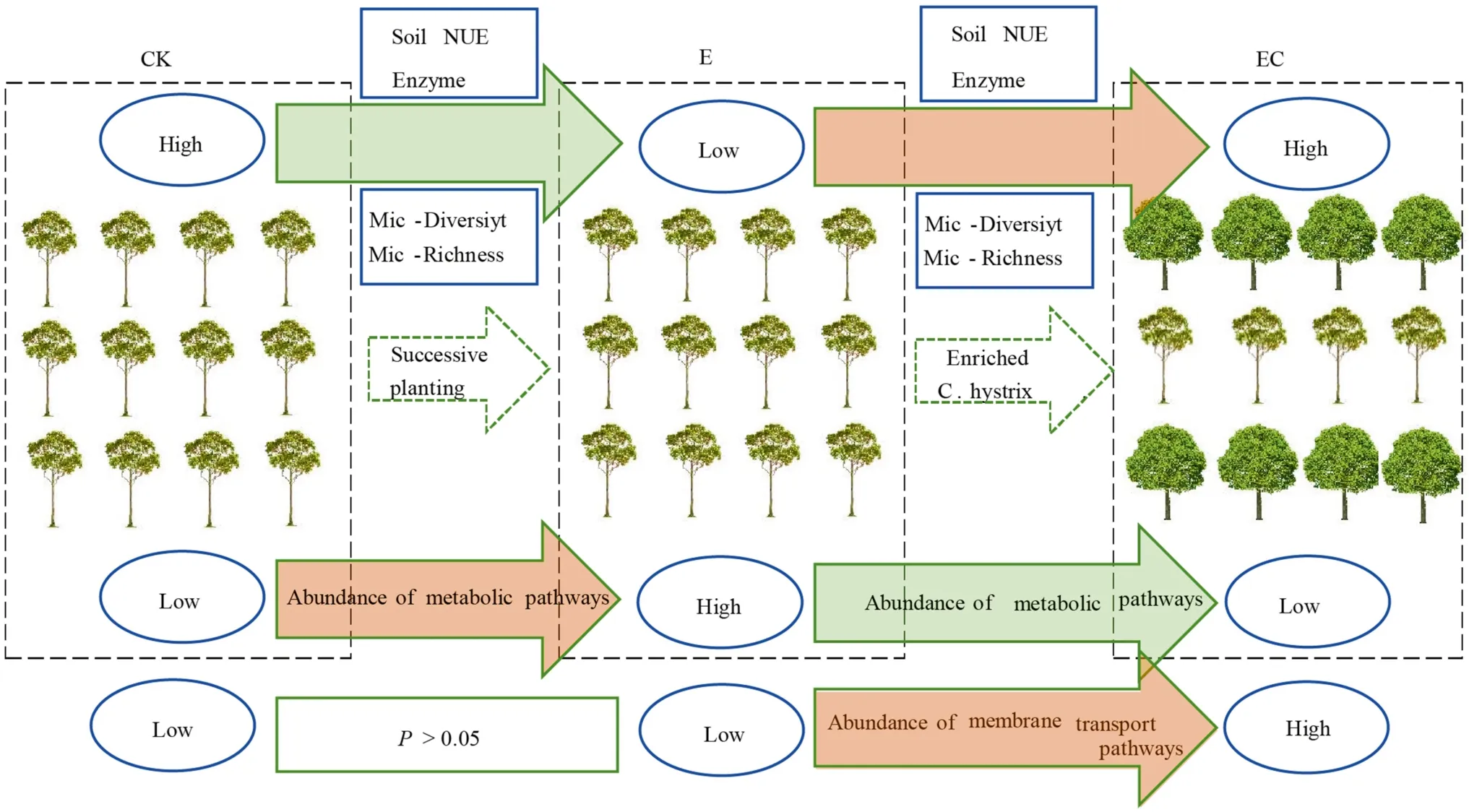
Fig.8 Conceptual models of soil biochemical properties under different Eucalyptus plantation management practices.CK–first rotation Eucalyptus plantation, E–third rotation Eucalyptus plantation,EC– E.urograndis with C.camphora, Soil NUE – soil nutrient status, Enzyme–soil enzyme activity, Mic-Diversity –microbial diversity, Mic-Richness–microbial richness
Soil invertin (INV), acid phosphatase (ACP), and urease activity (URE) are the key enzymes involved in soil organic C cycling, N, and P transformation, respectively.With these enzymes, C, N and P in organic matter are released and transported to plants or utilized by microorganisms (Gu et al.2009; Margalef et al.2017).Previous studies have shown that soil extracellular enzyme activity is positively related to organic matter concentration (Zhu et al.2020; Xu et al.2021), which is consistent with this study.Both the CK and EC plantings showed higher soil enzyme activity,which might be attributed to greater organic matter, total N,and available P concentrations to help form enzyme–humus complexes, thereby maintaining enzyme activity and delaying decomposition (Bastida et al.2012).The levels of OM,total N, and available P and the INV, ACP, and URE were also positively correlated with each other, confirming the coupling of soil C, N and P cycling.Similar results have been reported by Ren et al.( 2016) and Wang et al.( 2021).
Variations in soil bacterial community diversity
This study showed that the successive planting of monocultureEucalyptussignificantly reduced bacterial diversity and richness.Remarkably, the mixed reforestation ofEucalyptuswith native tree species increased bacterial diversity and richness, and for theE.urograndis×C.camphora(EC)planting, reached a significant level (Fig.8).This conclusion is consistent with previous research and may be attributed to the significant differences in soil chemical properties and enzyme activities caused by different composition of the plantations (Vos et al.2013; Pereira et al.2019; Zhu et al.2019).Rousk et al.( 2010) showed that soil bacterial diversity and richness were closely related to pH, and there was a positive correlation at pH 4–7.This is consistent with the results of this study, i.e., bacterial α-diversity indicators were positively correlated with pH, total N, and available P concentrations, supporting the importance of α-diversity in soil N, P dynamics (Arese et al.2020).In this study, continuous planting of the third-generation monocultureEucalyptus(E)increased acidification, while mixed plantings (especially EC) increased pH compared with E, providing a relatively suitable habitat for bacteria.On the other hand, increased litter diversity and dead roots under mixed plantings provides a good substrate for the accelerated decomposition of plant residues (Cizungu et al.2014), thus providing sufficient C and N sources for microbial growth (Demoling et al.2007),as well as a more diverse overall environment (Rottstock et al.2014).
Soil for E, EC, and EH were significantly lower than for CK, which may be because the first generationEucalyptusplantation was long undisturbed, and a fewAcaciaregenerated naturally in the plantation, which significantly improved C and N levels and N availability (Koutika et al.2014).However, there were no significant differences in, andamong the E, EC, and EH plantings,and the correlation with bacterial diversity was insignificant.This showed that, although new reforestation caused a significant decrease in soilconcentrations, bacterial diversity was not limited by N.Somewhat different from soil inorganic N, soil available P concentrations were significantly different among the four plantings and were positively correlated with bacterial diversity, indicating that soil bacteria at this development stage of the plantation were restricted by available P.
Variations in soil bacterial community structure
Although the composition of soil bacteria in theEucalyptusplantations and mixed plantations were similar, there were differences in community structure with the change in chemical properties.Diversity, acid phosphatase (ACP)activity, and OM levels were the most important factors for the bacterial community structure separation.Freedman and Zak ( 2015) showed that the distribution of dominant bacteria in soils of different types of plantations reflected the close relationship between soil spatial heterogeneity caused by environmental factors and soil microorganisms.Recent research showed that Chloroflexi, Acidobacteria, Proteobacteria, and Actinobacteria accounted for more than 80% of the total bacteria.Among them, Chloroflexi showed a significant negative correlation with bacterial diversity, ACP, and OM and were enriched in E and EH plantings, which may be partially related to Chloroflexi characteristics.Chloroflexi,widely known as oligotrophic bacteria, play key roles in photosynthesis, participating in the degradation of plantderived compounds (Krzmarzick et al.2012), and are more likely to survive in barren soils (Fierer et al.2007).However,Proteobacteria showed a positive correlation with the above key biochemical indexes, and were significantly enriched in the EC planting.This maybe because Proteobacteria, widely known as copiotrophic bacteria, grow rapidly in soils rich in unstable substrates (Newton and Mcmahon 2011; Wang et al.2017), and the increase in the abundance of Proteobacteria is beneficial to the decomposition of organic residues(Verzeaux et al.2016).
Among the top 10 dominant classes, AD3, Ktedonobacteria, and TK10 (Chloroflexi) were also significantly enriched in plantations E and EH, while Gammaproteobacteria, Deltaproteobacteria (Proteobacteria), and subgroup_6 (Acidobacteria) were higher in the controls and in the EC planting.The differential distribution of dominant classes reflects the positive response of soil bacterial community structure to nutrient differences caused by different forest management approaches.This result is consistent with previous studies that forest conversion significantly affects bacterial composition at the class level (Meng et al.2019).At the genus level,different plantation management approaches did not change the composition of the bacterial community (of the top 50 taxa), but there were significant differences in the relative abundance of dominant bacteria among the different plantations.This showed that bacterial communities adapted to the soil physicochemical changes by altering the abundance of dominant bacteria, reflecting the ecological adaptability of different types of bacteria.The genus-level bacterial communities of E and EH plantations were clustered, but different from the EC planting (consistent with PCoA results).This indicates that the effect of the EC planting on soil nutrients and bacterial community structure was significantly stronger than that of the EH planting in the early stage.
Variations in potential functions of the bacterial community
The metabolic function of soil bacteria changed significantly with soil chemical ( NO-3-N) and biochemical attributes (INV, Diversity, and URE).Cao et al.( 2020)indicated that soil prokaryotic microbial functions could be attributable to environmental selection for specific metabolic pathways based on soil physicochemical conditions.It is worth noting that several level-2 pathways related to metabolism were significantly enriched in E plantations, and were negatively correlated with INV,Diversity, and URE activity.Thismay be related to the decline of organic matter and nutrients such as total N and available P in the third generationEucalyptusplantation.These changes stimulated the coordinated regulation of numerous metabolic responses of microorganisms to adapt to the increasingly barren soil conditions (Song et al.2017).However, the mixed plantations enriched membrane transport-related genes (EC reached a significant level,p< 0.05).Do et al.( 2018) reported that most membrane transporters were present in bacterial cell membranes and were also involved in detoxification processes,organ growth, nutrition, development, and responses to abiotic and biotic stresses.Therefore, it may be inferred that the mixed plantations effectively alleviated the autotoxic effects caused by long-term continuous planting of a single species in early plantation stages (Zhang et al.2010).Although the benchmark results obtained showed good correlation with metagenome studies (Langille et al.2013), further experimental validation, such as shotgun metagenomics or targeted functional gene sequencing,should be performed.
Conclusion
This study revealed the effects of multi-generational planting of monocultureEucalyptusand a mixed approach ofEucalyptusand native species on soil biochemical properties and bacterial community structure and function.The pH,organic matter, nutrients (i.e., total N, NO-3-N, and available P), enzyme activities (i.e., INV, ACP, and URE), and bacterial α-diversity (i.e., diversity, richness, and evenness indexes) were significantly lower in third-generationEucalyptusplantations.However, the EC planting improved soil nutrient status, enzyme activity, and bacterial α-diversity,while the EH planting had no significant effect.Regarding soil bacterial community structure, although bacterial composition in monoculture plantations was similar to that in mixed plantations, LEFSE analysis showed that there were differences in the relative abundance of 26 bacterial clades among different plantation types, and that they were subordinate to Chloroflexi, Proteobacteria, and Acidobacteria.The oligotrophic bacteria Chloroflexi were significantly enriched in the third-generationEucalyptusplantation soil,while copiotrophic bacteria Proteobacteria increased significantly in theE.urograndiswithC.camphoraplantations.Tax4Fun prediction analysis showed that the relative abundance of multiple pathways related to metabolism increased significantly in the soil of the third generationEucalyptusplantations, while membrane transport-related genes were significantly enriched in plantations withC.camphora.This study suggests that continuous planting ofEucalyptusshould be avoided and it is recommended to create mixed plantations ofEucalyptusand native tree species on cutover areas that have experienced more than two generations of monoculture plantation management (Fig.8).
AcknowledgementsThe authors also appreciate the South China Experiment Nursery for support during the selection of suitable plots for this study.
Open AccessThis article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source,provide a link to the Creative Commons licence, and indicate if changes were made.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material.If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
 Journal of Forestry Research2022年4期
Journal of Forestry Research2022年4期
- Journal of Forestry Research的其它文章
- Journal of Forestry Research
- Reversibly photochromic wood constructed by depositing microencapsulated/polydimethylsiloxane composite coating
- Surveillance of pine wilt disease by high resolution satellite
- Adaptation of pine wood nematode, Bursap helenchus xylophilus,early in its interaction with two P inus species that differ in resistance
- Pine wilt disease detection in high-resolution UAV images using object-oriented classification
- Transcriptome analysis shows nicotinamide seed treatment alters expression of genes involved in defense and epigenetic processes in roots of seedlings of Picea abies
