p62/SQSTM1 Participates in the Innate Immune Response of Macrophages Against Candida albicans Infection
Yan-Zhi He, Zhi-Min Duan, Xu Chen,3,*, Min Li,3,*
1Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu 211166, China; 2Department of Dermatology, Hospital for Skin Diseases (Institute of Dermatology), Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, Jiangsu 210042, China; 3Jiangsu Key Laboratory of Molecular Biology for Skin Diseases and STIs,Hospital for Skin Diseases (Institute of Dermatology), Chinese Academy of Medical Sciences and Peking Union Medical College,Nanjing, Jiangsu 210042, China.
Abstract
Keywords: p62/SQSTM1, Candida albicans, macrophage, innate immune response, phagocytosis
Introduction
During the past few decades,infection by C. albicans has emerged as a crucial public health problem in parallel with the increased population of individuals with immune suppression.1C. albicans can infect local tissues (eg, skin and vagina) and multiple organ systems, and systemic infection leads to high morbidity and mortality in hospitalized patients.2-3The innate immune response plays a key role in the body’s defense against the challenge of C.albicans.As important members of the innate immune cell population, macrophages recognize and phagocytize C.albicans,thus functioning in a key defensive role.
Autophagy is an essential self-digestive cellular mechanism.4Recent reports have indicated that autophagy is involved in the clearance of invading microorganisms such as bacteria, viruses, and fungi.5Several sequestosome 1-like receptors such as SQSTM1(also known as p62)and neighbor of BRCA1 gene 1 have been found to participate the machinery of selective autophagy.6During the process of selective autophagy, sequestosome 1-like receptors connect the microtubule-associated protein 1 light chain 3 and autophagic cargo for the initiation of autophagydepen-dent degradation after fusion of autophagosomes and lysosomes.7
p62 functions as a stress-inducible scaffold protein that selects and tags specific components in the cytoplasm for sequestration into autophagosomes.8Accordingly, p62 plays essential roles in cell biology, including energy regulation,differentiation,and proliferation.p62 reportedly contributes to a wide range of human disorders such as bone and muscle diseases, neurodegeneration, metabolic diseases,and some types of cancer.9p62 is also required for the autophagic host defense against mycobacterial challenges.p62 loss-of-function mutation impairs the autophagic targeting function against mycobacteria,and p62 gene knock-out increases the susceptibility of zebra-fish to mycobacterial infection.10Zhang et al.11reported that p62 participates in recognizing Listeria, Salmonella, and Shigella and contributes to the elimination of these pathogens through autophagy initiation. However, the mechanism underlying how p62 participates in host defense of macrophages against fungal infections remains unclear.
In this study, we investigated the role of p62 in the immune response of cultured primary murine macrophages stimulated by C.albicans in vitro.Our data show that p62 contributes to phagocytosis and some crucial immune responses of macrophages after stimulation by C.albicans.
Materials and methods
C. albicans strains
The SC5314 strain of C. albicans used in this study was kept at the China Center for Medical Microorganisms Culture Collection,Hospital for Skin Diseases(Institute of Dermatology), Chinese Academy of Medical Sciences,and Peking Union Medical College, Nanjing, China. C.albicans was stored in 30% glycerol solution at a temperature of -80°C, and maintained in yeast extract peptone dextrose agar medium containing glucose (2%),peptone(2%),and yeast extract(1%)at a temperature of 4°C. For in vitro assays, the C. albicans was cultured in yeast extract peptone dextrose agar medium at 28°C overnight and maintained at a speed of 2.655g to obtain the yeast form of C.albicans.We counted the yeast forms of C.albicans using a hemocytometer and normalized the C. albicans for the appropriate fungal concentrations.
Isolation and culture of bone marrow-derived macrophages
Weharvestedcellsfromthebonemarrowof8-to10-week-old C57BL/6 mice(GemPharmatech Co.,Ltd.,Nanjing,China).We flushed the femurs and tibias of the mice with phosphatebuffered saline,and the harvested cells were then cultured in differentiation medium(Dulbecco minimum essential medium)containing 50ng/mL mouse macrophage colony-stimulating factor(Miltenyi Biotec,Bergisch Gladbach,Germany),10%fetal bovine serum,100μg/mL streptomycin solution,and 100 IU/mL penicillin(both from Hyclone Laboratories,Logan, UT, USA) for 7days. We replaced the cultured differentiation medium every 3days. Six-well tissue culture plates were used to culture the bone marrow-derived macrophages(BMDMs)(2×106cells per well).
Antibodies
The primary antibodies used in this study were antiglyceraldehyde 3-phosphate dehydrogenase (#2118; Cell Signaling Technology, Danvers, MA, USA) and anti-p62(#109012;Abcam,Cambridge,MA,USA).The secondary antibody used in the study was anti-rabbit immunoglobulin G (horseradish peroxidase-linked) (#7074; Cell Signaling Technology).
Western blotting
We lysed the BMDMs in RIPA Lysis Buffer containing a protease inhibitor cocktail(Roche Applied Science,Basel,Switzerland) and the phosphatase inhibitor PhosSTOP(Roche Applied Science).A BCA assay kit(Thermo Fisher Scientific,Waltham,MA,USA)was used to determine the concentration of protein in the samples.Equal amounts of each total protein sample were loaded into sodium dodecyl sulfate-polyacrylamide gels (4%-12%; Thermo Fisher Scientific). We transferred the protein from the gels to polyvinylidene fluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA, USA), and the PVDF membranes were blocked in EveryBlot Blocking Buffer(Bio-Rad Laboratories).Next,the PVDF membranes were incubated with the indicated primary and secondary antibodies.We imaged the indicated protein band by chemiluminescence.Quantity One software(Bio-Rad Laboratories)was used to quantify the intensity of the indicated protein bands. We selected glyceraldehyde 3-phosphate dehydrogenase as the loading control in this study.
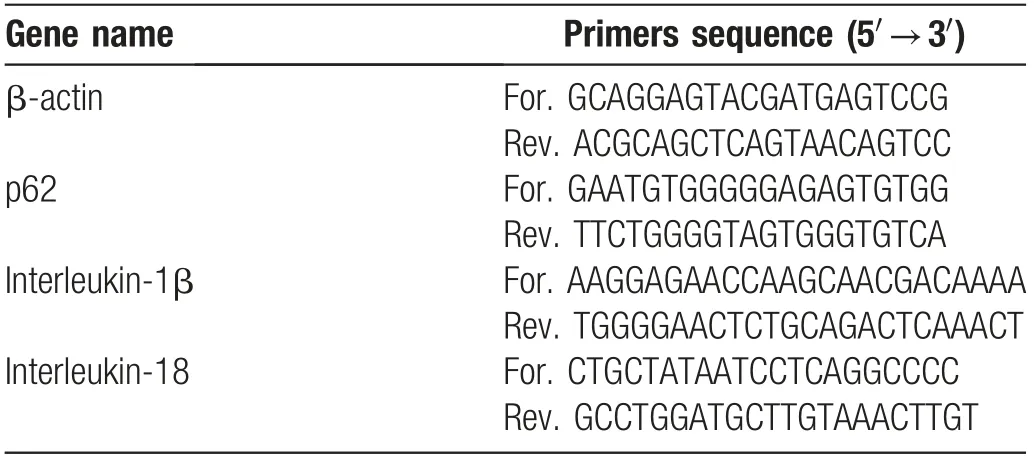
Table 1 Names and sequences of the primers of four target genes.
Quantitative reverse transcription PCR assay
We extracted the total RNA from BMDMs using an RNAsimple Total RNA Kit (Tiangen Biotech, Beijing,China). The samples were then reverse-transcribed to complementary DNA by the PrimeScriptTMRT reagent Kit (RR036A-1; Takara, Dalian, China) according to the manufacturer’s protocol. We performed the quantitative reverse transcription (qRT)-PCR using an instrument(LightCycler?480InstrumentII;RocheDiagnostics).β-actin served as the housekeeping gene. The primer sequences of the specific genes used in qRT-PCR are listed in Table 1.
Transfection of small interfering RNA
We transfected 100nmol/L of p62 small interfering RNA(siRNA)(GGACCCAUCUGUCUUCAAATT,AAAGUGUCCAUGUUUCAGCTT)and control siRNA(UUCUCCGAACGUGUCACGUTT, ACGUGACACGUUCGGAGAATT)(GenePharma,Shanghai,China)into the BMDMs using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Immunofluorescence assays
Cells were transfected using Lab-Tek II CC2TM(Thermo Fisher Scientific)at a density of 4×105cells/well for at least 24hours. The C. albicans yeast form, which was already marked by Uvitex 2B dye(Polysciences,Inc.,Warrington,PA,USA),was added to the cultured medium for 8hours.p62 and nucleus were marked by Alexa Fluor 568 antimouse F4/80 antibody (BioLegend, San Diego, CA, USA)and SYTOX Green Nucleic Acid Stain (Thermo Fisher Scientific). LysoTracker Probes (L12492; Thermo Fisher Scientific)were added to the culture medium at 50nmol/L,and the cells were incubated for 30 minutes at 37°C(λex=647nm and λem=668nm). Cells were imaged using an Olympus FV1000 laser scanning confocal microscope(Olympus,Tokyo,Japan)(Uvitex 2B scanning,λex=350 nm and λem=435nm;SYTOX Green scanning,λex=504 nm and λem=523nm).
Phagocytosis assay
BMDMs were seeded in 12-well plates at a density of 4×105cells/well. The C. albicans yeast form, which was already marked by FUN-1(Sigma-Aldrich,St.Louis,MO,USA),was added to the cultured medium 2hours after cell seeding at a specific ratio(BMDM:yeast=1:3).At 0.5,1,and 2hours after challenge by C. albicans yeasts, the BMDMs were washed twice with phosphate-buffered saline.We then incubated the BMDMs with Alexa Fluor 647 anti-mouse F4/80 antibody(BioLegend)according to the manufacturer’s instructions.The samples of BMDMs were analyzed by flow cytometry.
Reactive oxygen species assay
The level of reactive oxygen species(ROS)was measured by an ROS detection kit (CellROXTMOxidative Stress Reagent C10422;Thermo Fisher Scientific).The BMDMs were seeded in a 12-well plate. After treatment with C.albicans, the BMDMs were incubated with 5μmol/L of CellROXTMat 4°C for 20 minutes. The BMDMs were then incubated with Alexa Fluor 488 anti-mouse F4/80 antibody (BioLegend) and propidium iodide (Thermo Fisher Scientific)according to the manufacturer’s instructions. The samples were analyzed by flow cytometry.
Statistical analysis
Independent experiments were replicated at least three times.Statistical analyses were performed with GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA,USA).Univariate analysis of variance or Student t test was used to analyze the data. Data are shown as mean±standard deviation,and a P value of <0.05 was considered statistically significant.
Results
C. albicans increased the expression of p62 at themRNA and protein levels in BMDMs
The relative level of p62 mRNA was upregulated in a dose-dependent manner in the BMDMs challenged by C.albicans. The relative levels of p62 mRNA at different ratios (BMDMs:C. albicans) were 1.893±0.2156 (1:1,P<0.05), 2.873±0.4787 (1:3, P<0.05), and 3.556±0.2892(1:5,P<0.01)(Fig.1A).The protein level of p62 was also increased in BMDMs after treatment with C.albicans at different ratios (BMDMs:C. albicans)(Fig. 1B). We marked the lysosomes with LysoTracker probes,which exhibited red fluorescence,and the p62 and C. albicans were also visualized by immunofluorescence.We found that p62 was recruited together with the phagocy-tized C. albicans and lysosomes (Fig. 1C).
Decrease in p62 expression reduced the phagocytosis rate of C. albicans in BMDMs
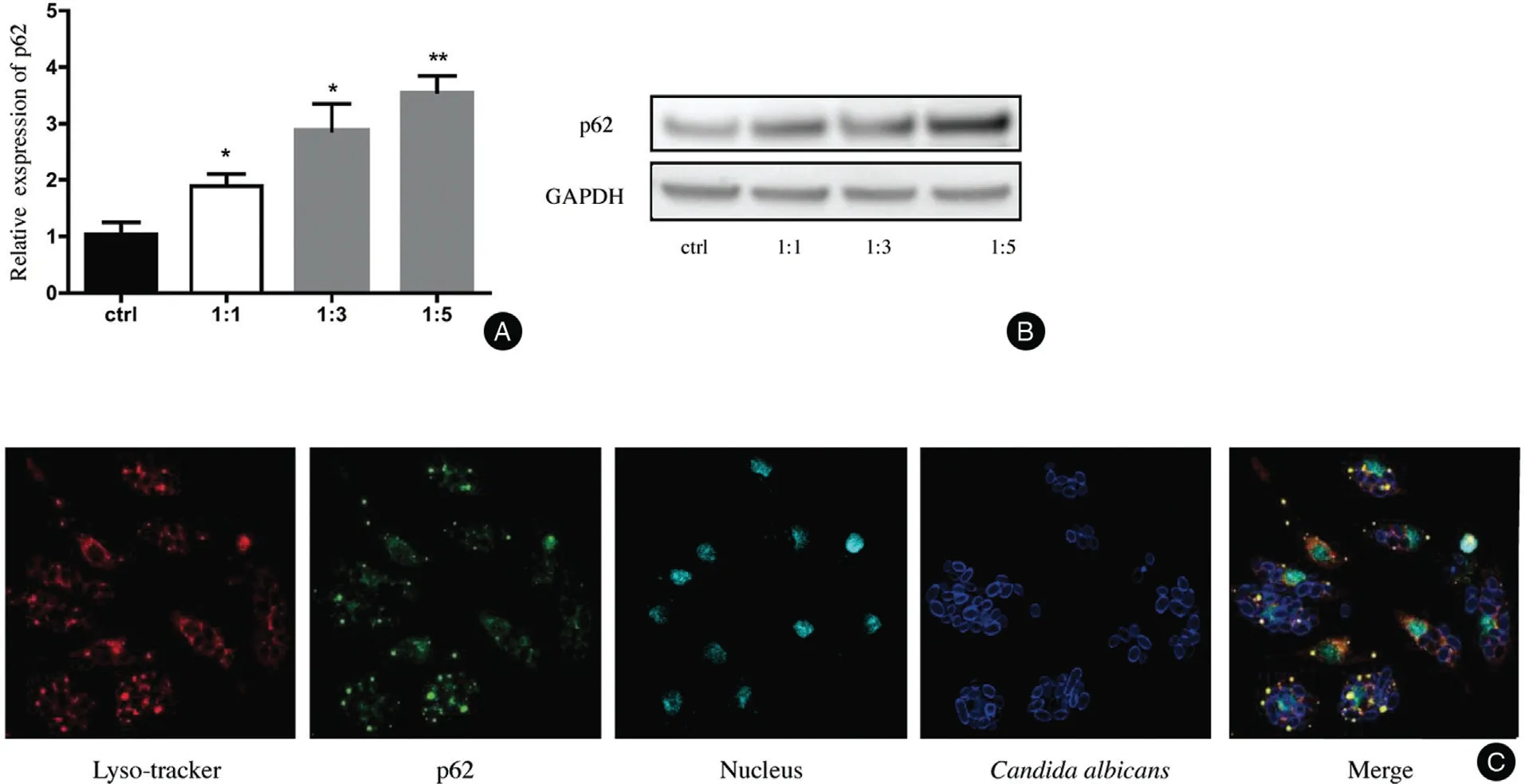
Figure 1. C.albicans increased the expression of p62 in BMDMs.(A and B):Treatment by C.albicans for 4hours induced an increase in p62 at both the mRNA(A)and protein(B)levels in bone marrow-derived macrophages compared with the control.The ratios of cells vs.yeast were 1:1, 1:3, and 1:5, respectively. (C) The C. albicans was labeled with Uvitex 2B, and p62 and LysoTracker were co-localized around the C.albicans.The mean±standard deviation of data from three independent experiments is shown.*P<0.05,**P<0.01.C. albicans: Candida albicans; ctrl: control; GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
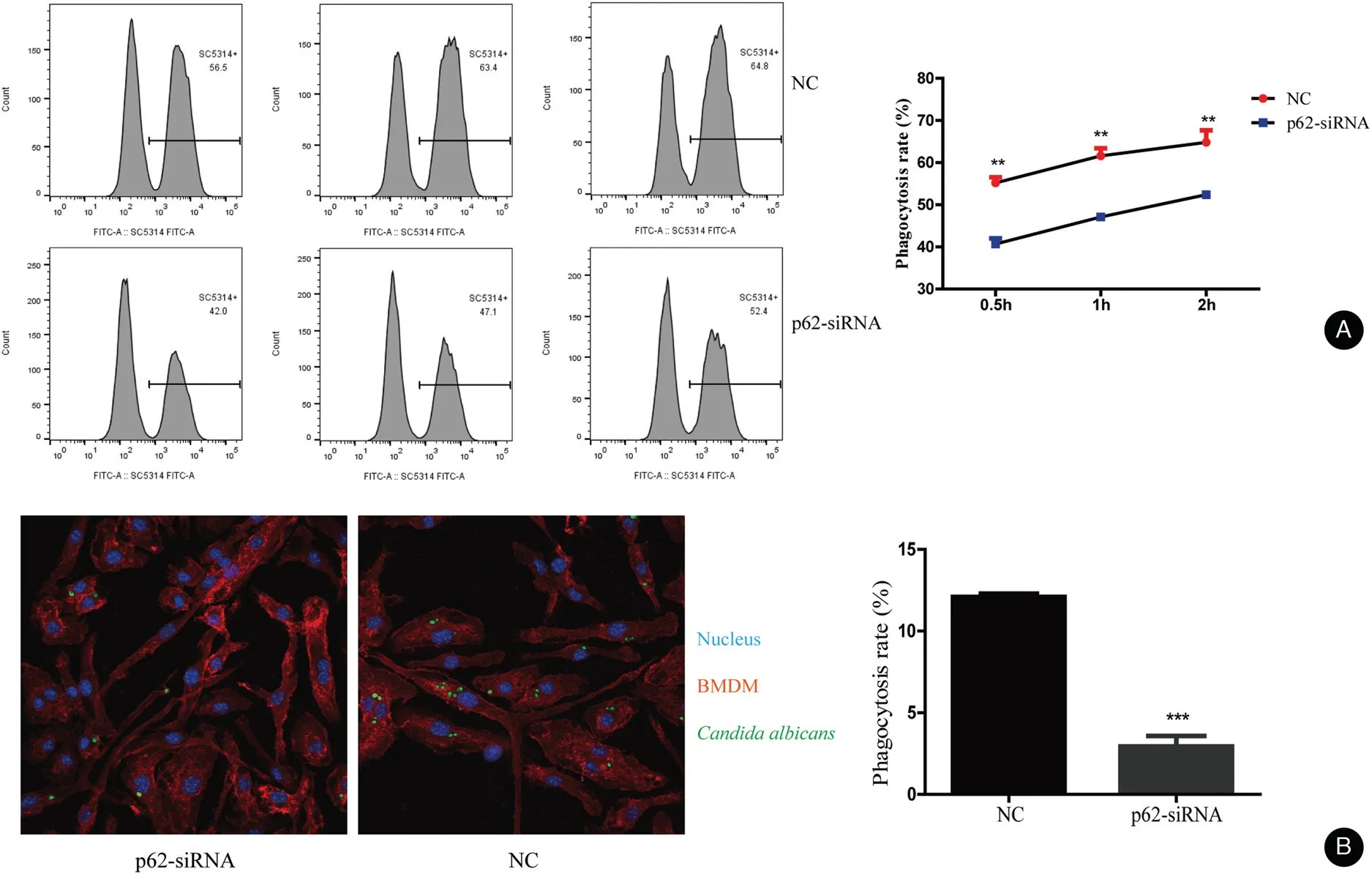
Figure 2. p62 influence the phagocytosis rate of Candida albicans in BMDMs.(A)The phagocytic activity of BMDMs was measured by flow cytometry after treatment at a 1:5 ratio of cells vs yeast for 0.5,1,and 2hours,respectively.The mean±standard deviation of data from three independent experiments is shown.(B)The rate of phagocytosis of C.albicans by BMBMs transfected with p62 siRNAs was lower than that of the NC as observed by a laser scanning confocal microscope,BMDMs marked by Alexa Fluor 647 anti-mouse F4/80 antibody,and nucleus marked by Hoechst.**P<0.01. BMDMs: bone marrow-derived macrophages; NC: negative control.
We knocked down the p62 expression by transfection of p62 siRNA into BMDMs.We monitored the knock-down efficiency by qRT-PCR and western blotting (Fig. S1,http://links.lww.com/JD9/A23). The phagocytosis results measured by flow cytometry are shown in Figure 2A.The rate of phagocytosis was lower in the p62 siRNA group than that in the negative control(NC)group.The rate of phagocytosis in the NC group was 55.23%±0.72%,60.80%±1.78%, and 64.43%±2.00% at 0.5, 1, and 2 hours,respectively. However, the rate of phagocytosis in the p62 siRNA group was 39.70%±1.69%, 46.70%±0.89%, and 51.90%±0.98% at 0.5, 1, and 2hours,respectively. Moreover, the images taken by the laser scanning confocal microscope showed that the level of phagocytosed C. albicans in BMDMs was lower in BMDMs transfected with p62 siRNA than in NCs(BMDMs transfected with control siRNA) (Fig. 2B).
Decrease in p62 expression reduced the ROS levels in BMDMs stimulated by C. albicans
We found that the levels of ROS production were lower in BMDMs transfected with p62 siRNA than in the NC group.The ROS levels in BMDMs of the NC group were 13,430±1,860, 13,690±2,611, and 17,720±1,814 at 0.5, 1, and 2hours after treatment of C. albicans,respectively. However, the ROS levels in BMDMs transfected with p62 siRNA were 4,269±393, 4,967±721,and 7,647±1,950 at 0.5, 1, and 2hours, respectively(Fig. 3). At the three time points after treatment with C.albicans, the ROS levels were decreased in BMDMs transfected with p62 siRNA.
p62 knock-down decreased the mRNA levels of proinflammatory cytokines interleukin (IL)-1β and IL-18 in BMDMs challenged by C. albicans
We found that the mRNA levels of IL-1β and IL-18 were lower in BMDMs transfected with p62 siRNA than in NCs after treatment with C.albicans(Fig.4).The mRNA level of IL-1β in NCs was 12.12±0.54 and 26.2±4.67 at 4 and 12hours, respectively, whereas that in BMDMs trans-fected with p62 siRNA was 6.14±1.63 and 8.81±0.86 at 4 and 12hours,respectively.We observed similar results in the assay of IL-18 mRNA. The levels of IL-18 mRNA at 4 and 12hours were higher in the NCs(0.97±0.06 and 2.23±0.46, respectively) than in BMDMs treated with p62 siRNA (0.38±0.02 and 0.44±0.02,respectively; P<0.05 for both time points).
Discussion
In the present study, we found that p62 can colocalize with C. albicans. We also demonstrated that p62 may participate in the phagocytosis of C.albicans and promote the innate immune response of macrophages.
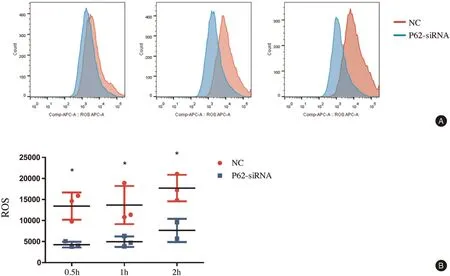
Figure 3. p62 influenced the production of ROS in BMDMs after stimulation by Candida albicans detected by flow cytometry. A: the production of ROS was decreased by p62 siRNA.The ratio of cells vs yeast was 1:5,and co-culture was performed for 0.5,1,and 2hours.B:The mean±standard deviation of data from three independent experiments is shown.*P<0.05. BMDMs: bone marrow-derived macrophages; NC: negative control; ROS: reactive oxygen species.
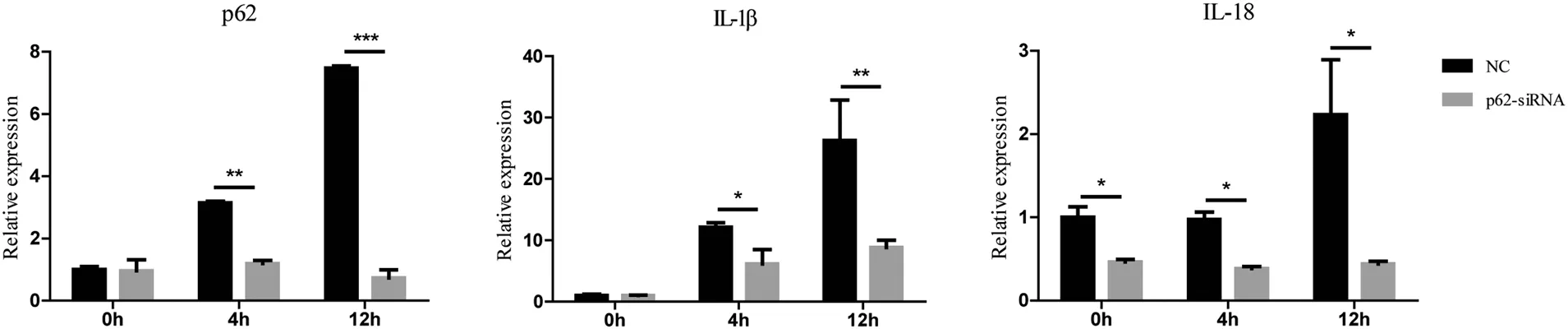
Figure 4. Effects of p62 on the relative mRNA expression of proinflammatory cytokines in BMDMs induced by Candida albicans. After treatment with Candida albicans,the mRNA levels of IL-1 β and IL-18 were lower in BMDMs transfec*ed with p62 siRNA than in the NC.All values are presented as mean±standard deviation from three independent experiments.*P<0.05,**P<0.01,***P<0.001.BMDMs:bone marrow-derived macrophages; IL: interleukin; NC: negative control.
The function of macrophages mainly depends on recognition and phagocytosis in the immune response to a challenge by C. albicans.12β-glucan (particularly β-(1,3)-glucan) is a core pathogen-associated molecular pattern(PAMP)of C.albicans.13This PAMP is recognized by dectin-1 (a C-type lectin-like receptor), and its activation leads to the induction of phagocytosis of invading fungal pathogens.14C.albicans reportedly induces the upregu-lation of Sel1(a small secreted cysteine-rich protein). Moreover, Sel1 can lead to activation of the mitogen-activated protein kinase and nuclear factor-κB(NF-κB) signaling pathways, and it induces the production of chemokines and proinflammatory cytokines to antagonize the C.albicans infection.15Additionally,ROS production in phagocytes is a crucial cellular mechanism in defending against fungal invasion by the host defense system.16ROS serve as messenger molecules that participate in the innate immune response.For example,NLRP3 inflammasome (cryopyrin) is activated by the increase in ROS production during defense against the invasion of pathogens.17-18
In this study, we found that the phagocytic ability and ROS production were decreased in BMDMs with knockdown of p62 after the challenge by C.albicans,indicating that p62 promotes the phagocytosis of C. albicans. Our findings are consistent with the results of a study by Bonilla et al.,19who found that the increase in scavenger receptors resulting from the accumulation of p62 in Atg7-/-macrophages facilitated phagocytosis against the challenge byM.tuberculosis.However,whetherp62isinvolvedinthe recognition of C.albicans by the classic receptors(dectin-1 or TLR2/4)for PAMPs remains to be investigated.
Our study also showed that the increase in IL-1β and IL-18 mRNA in macrophages challenged by C.albicans was inhibited by the interference of p62 through siRNA transfection, indicating that p62 is conducive to the production of some proinflammatory cytokines in macrophages during the immune response against C. albicans.p62 can reportedly activate NF-κB signaling through tumor necrosis factor receptor-associated factor 6 and thereby enhance the inflammatory response.8Therefore,we speculated that the effects of p62 in promotion of the inflammatory response might depend on assistance by the NF-κB pathway.
Overall,our study showed that challenge by C.albicans induces an increase in p62 expression at both the mRNA level and protein level in macrophages in vitro. Importantly,p62 protein aggregates around the phagocytized C.albicans spores in granular form,and knock-down of p62 inhibits phagocytic ability,ROS production,and upregulation of proinflammatory factors IL-1β and IL-18.
However,how C.albicans upregulates the p62 protein remains unclear. It has been reported that the phosphorylation of p62 at the Ser403 site strongly increases the affinity of the ubiquitin-associated domain in p62 to the ubiquitin chains linked with K48 and K63 and facilitates the autophagic clearance to p62 and the protein of polyubiquitination.20Whether this mechanism is involved in the removal of C.albicans needs to be verified by further experiments.
In this study,we elucidated the correlation between p62 protein and the innate immune response in macrophages.Regulation targeted to p62 might be considered as a potential strategy to improve the prognosis of C.albicans in macrophages and strengthen the role of p62 against C.albicans infection.However,this study did not analyze the mechanisms and pathways that p62 relies on to recognize and phagocytose C. albicans, which is the limitation of this study.More insight is needed into the mechanism by which p62 contributes to the recognition, phagocytosis,and further immune responses in macrophages against challenge by C. albicans.
Source of funding
The study was supported by the National Natural Science Foundation of China (Nos. 82103749, 81773338, and 82173432),ChineseAcademyofMedicalSciencesMedicine andHealthTechnologyInnovationProject(No.2017-I2M-1-017), Natural Science Foundation of Jiangsu Province(No.BK20190144),and the Nanjing Incubation Program for National Clinical Research Center(No.2019060001).
- 國際皮膚性病學(xué)雜志的其它文章
- Clinical Features and Corrected Factors with Neurosyphilis in HIV/Syphilis Co-Infected Patients Based on Stage of Syphilis
- Dermoscopic Features of Basal Cell Carcinoma and Their Association with Histological Types in A Chinese Population: A Perspective Study
- Frostbite in Southwest China: A Single-Center Retrospective Analysis
- Role of Epigenetics in the Pathogenesis of Systemic Sclerosis
- Skin Organoid Research Progress and Potential Applications
- Dermatoscopy in the Diagnosis of Vulvar Basal Cell Carcinoma: A Case Report

