Bioinformatics analysis of structure and function in the MRP gene family and its expression in response to various drugs in Bursaphelenchus xylophilus
Jian Diao · Xin Hao · Wei Ma · Ling Ma
Abstract Genes homologous to members of the MRP gene family in Caenorhabditis elegans are important in drug resistance. To further explore the molecular mechanism of drug resistance in pine wood nematode ( Bursaphelenchus xylophilus), we used bioinformatics approaches to analyze genomic data for B. xylophilus and identified Bx- MRP genes. We predicted the structure and function of the genes and encoded proteins. Using bioinformatics programs to predict and analyze various properties of the predicted proteins, including hydrophobicity, transmembrane regions, phosphorylation sites, and topologically isomeric structures, of these Bx- MRP genes, we determined that they function in transmembrane transport. From the results of RT-qPCR, the Bx- MRP family members confer significant differential resistance to different drug treatments. After treatment with different concentrations ofemamectin benzoate, avermectin and matrine, the expression ofeach gene increased with increasing drug concentrations, indicating that the family members play a positive role in the regulation of multidrug resistance.
Keywords Bursaphelenchus xylophilus · Bx- MRP gene family · Gene and protein structure · Bioinformatics · Multidrug stress
Introduction
As one of the most important wood source species on the planet, pine has been widely recognized for its ecological benefits, social benefits and economic importance. Long an important part of the human lifestyle, they are also typical afforestation species in China (Soliman et al. 2012; Futai 2013). However, pine trees are highly susceptible to pine wood nematode disease (PWD) in nature (Li et al. 2019a, b), and Bursaphelenchus xylophilus has also become a major threat to pine afforestation (Mota and Vieira 2008). Pine wilt disease (PWD) caused by the highly virulent, migratory, facultative endoparasite B. xylophilus (Liu et al. 2019a, b), has now caused extremely severe damage to forest ecosystems (Liu et al. 2019a, b). The prevention and control of PWD is mainly achieved with aerial application of synthetic insecticides, fumigation of infected trees, manual removal of contaminated pine forests and application of natural enemies (Kwon 2008). The use of synthetic pesticides, however, will impact environmental and human health and damage nontarget organisms (Zhao 2008). At present, a safer method of prevention and control is to protect pine trees from nematode infection by injecting plant nematicides. Two nematicidal avermectin compounds (abamectin and emamectin benzoate) for the prevention of PWD have been widely used.
The ATP-binding cassette (ABC) transporter is the most conserved protein superfamily, expressed from eukaryotes to vertebrates. Because of the high diversity in their expression levels, members of the ABC transporter family play a vital role in all organisms in nature (Domenichini et al. 2019). Since the discovery of the MDR1 gene (Yang et al. 2002), this family has been recognized as the major protein conferring multidrug resistance. The main members involved in multidrug resistance are ABCB, ABCC, ABCG and other subfamilies, and they act synergistically in the multidrug resistance process. There is no correlation between the expression of different members, and the substrates for these members can overlap. In this review, we discuss genes that belong to the ABCC subfamily members. Due to the important role of this subfamily member in multidrug resistance and drug effl ux, members of the ABCC subfamily are often referred to as multidrug resistance proteins (MRPs).
According to previous studies, the MRP in the ABC transporter family is not only an important related protein in the nematode resistance metabolic system, but also an important drug-sensitive target (Hyde et al. 1990). Because of the other characteristics of the subfamily, MRP proteins are potential targets for the control of pine wilt disease. This study also has extremely important guiding significance in the control of B. xylophilus (Kikuchi et al. 2011). At present, research on MRP genes has mainly focused on nematodes of animals. Among the plant parasitic nematodes, only our research group has studied the role of Bx- mrp1 in resistance (Hao et al. 2018). Considering that resistance and metabolic mechanisms in plant parasitic nematodes are the main ways to tolerate drugs, the anti-drug function of the mrp subfamily in B. xylophilus was studied in depth to provide a theoretical basis for the development of new nematode pesticide synergists and control technologies (Hao et al. 2018). Genes in the B. xylophilus resistance gene family Bx- mrp were identified, and their protein structure and function were analyzed using bioinformatics.
Materials and methods
Nematode culture and RNA extraction after drug treatment
Bursaphelenchus xylophilus, maintained in the Forestry Protection Laboratory of Northeast Forestry University, Harbin, China, were kindly provided by the Chinese Academy of Forestry, Beijing, China. The pine wood nematode was cultured in the dark on Botrytis cinerea (Wang et al. 2019a, b) at 25 °C for 5–7 days. Healthy pine wood nematodes were then selected and treated with the following drug solutions at 25 °C for 12 h in the dark: emamectin benzoate solution (Hubei Jusheng, CAS No.: 155569-91-8) in DEPC-H2O at 0, 50, 100, 150, 200 μg mL; avermectin solution (Shanghai Jizhi, CAS No.: 71751-41-2) in DEPC-H2O at 0, 100, 150, 200, 250 μg mL?1; matrine solution (Shanghai Yuanye, CAS No.: 519-02-8) in DEPC-H2O at 0, 2, 4, 6, 8, 10 μg mL?1(Wang et al. 2019a, b). The nematodes (mixed insect age 100 μL) were washed with M9 buffer, centrifuged, and the supernatant was removed. After liquid nitrogen milling of the nematodes, 100 μL of TRIzol (Invitrogen, USA) was added to extract total RNA, which was then reverse transcribed into cDNA for subsequent experiments (Wang et al. 2012, 2017).
Phylogenetic relationship and physicochemical properties of the Bx- mrp gene family
We downloaded the amino acid sequences from the NCBI website ( https://www.ncbi.nlm.nih.gov/). The amino acid sequences were aligned using MEGA 5.1 software (Tamura et al. 2011), and a phylogenetic tree was constructed using the neighbor-joining (NJ) method, 1000 iterations of bootstrap resamples and the Poisson model.
To further verify the evolutionary relationship ofeach protein, we constructed the phylogenetic tree using the maximum likelihood (ML) method (Tamura et al. 2011). We searched for DNA-binding domain sequences among the Cbr- mrp family in the Pfam database ( http://pfam.xfam.org/) (Finn et al. 2016) and aligned the sequences obtained. The DNA binding domain was then visually compared via WebLogo ( http://weblo go.berke ley.edu/logo.cgi) (Crooks et al. 2004). The ProtParam ( https ://web.expas y.org/protp aram/) function (Wilkins et al. 1999) can be used to predict the physical and chemical properties of proteins such as number of amino acids, molecular weight, theoretical pI, aliphatic index, grand average of hydropathicity, formula, total number of atoms and Instability index.
Protein sequence motif analysis
The protein sequence motifs can be predicted by the MEME method ( http://meme-suite.org/) (Bailey et al. 2015), then the motifs can be identified using Evolview ( http://www.evolg enius.info/evolv iew) (Zhang et al. 2012; He et al. 2016). The motifs were annotated using the Pfam database ( http://pfam.xfam.org/) (Finn et al. 2016).
Hydrophobicity and transmembrane helical segments analysis
The hydrophobicity of the protein was analyzed using the ExPASy ProtScale website ( https ://web.expas y.org/prots cale/) (Seddigh 2017; Seddigh and Darabi 2018). TMHMM ( http://www.cbs.dtu.dk/servi ces/TMHMM/) (Krogh et al. 2001) was used to predict transmembrane helices in proteins.
Phosphorylation sites prediction
The Netphos 3.1 website ( http://www.cbs.dtu.dk/servi ces/NetPh os/) (Ma et al. 2018) was used to predict phosphorylation sites.
Topological heterogeneity model prediction
The topological heterogeneity model of the protein was analyzed using the Protter website ( http://wlab.ethz.ch/prott er/start/) (Omasits et al. 2014).
Verification of predictions using reverse-transcription quantitative real-time PCR
For molecular validation ofour bioinformatics results using reverse-transcription quantitative real-time PCR (RT-qPCR) on 11 genes studied here, we used 28S and β-actin reference genes (Wang et al. 2017) and respective primer pairs 28S-F (GTG CGT ATT CAG CCT TCT GG) and 28S-R (AAC CGA ACA CGC GAC AAT AG); β-actin-F (TTG GCT GGC CGT GAC TTG AC) and β-actin-R (GCG GTG GCC ATC TCC TGT TC). The other primer sequences are listed in Table S1. The expression level ofeach gene was calculated as relative to the expression of the corresponding gene in the untreated nematode.
Results
Identification of Bx- mrp gene family and analysis of its physicochemical properties
We identified 10 Bx- mrp gene members of B. xylophilus from the genome and named the genes from Bx- mrp1 to Bx- mrp10 (Table S2). Members of this gene family were predicted to have the same DNA-binding domain. The results of sequence alignment of the DNA binding domain about the Bx- mrp gene family of B. xylophilus are listed in Table S3, and Fig.1 shows the DNA-binding domain alignment logo of 155 amino acids. The physicochemical analyses showed that the length and molecular weight of the 10 corresponding proteins differed significantly (Table S2). The average length of amino acids is 975.1, ranging from 342 to 1552. The average molecular weight is 108,924.287 Da (38,060.38–174,290.56 Da). The theoretical isoelectric points of these proteins ranges from 4.86 to 9.01. The aliphatic index varies between 90.86 and 109.93. It can be seen from the data that the thermal stability of these proteins has changed significantly. The average value of protein hydropathicity varied from ? 0.282 to 0.157, except for Bx- mrp8 and Bx- mrp10, other values were predicted to be positive, indicating that Bx- mrp8 and Bx- mrp10 are hydrophilic proteins, and the other eight are hydrophobic proteins. These proteins consist of five atoms, which are C, H, N, O and S. The total number of atoms ranges from 5385 to 24,690. Details are shown in Table S2. The protein instability index ranged from 29.65 to 42.51, and the Bx- mrp1, Bx- mrp2, Bx- mrp4, Bx- mrp6, Bx- mrp7 and Bx- mrp9 indices were all less than 40, indicating that the protein structure was stable. The indices for the other proteins were over 40, indicating that their structures were not stable.
Phylogenetic trees and protein sequence motif analyses
To reveal the relationship between members of the Bx- mrp gene family in B. xylophilus, we constructed an NJ-phylogenetic tree using their protein sequences (Fig.S1). Use the bootstrap resample test as the reliability basis for the phylogenetic tree results (Fig.S1). To further verify the reliability of the NJ-phylogenetic tree, we constructed the ML-phylogenetic tree using the best model of JTT + G + F (Fig.S2). The consistency of the two phylogenetic tree results is very good.
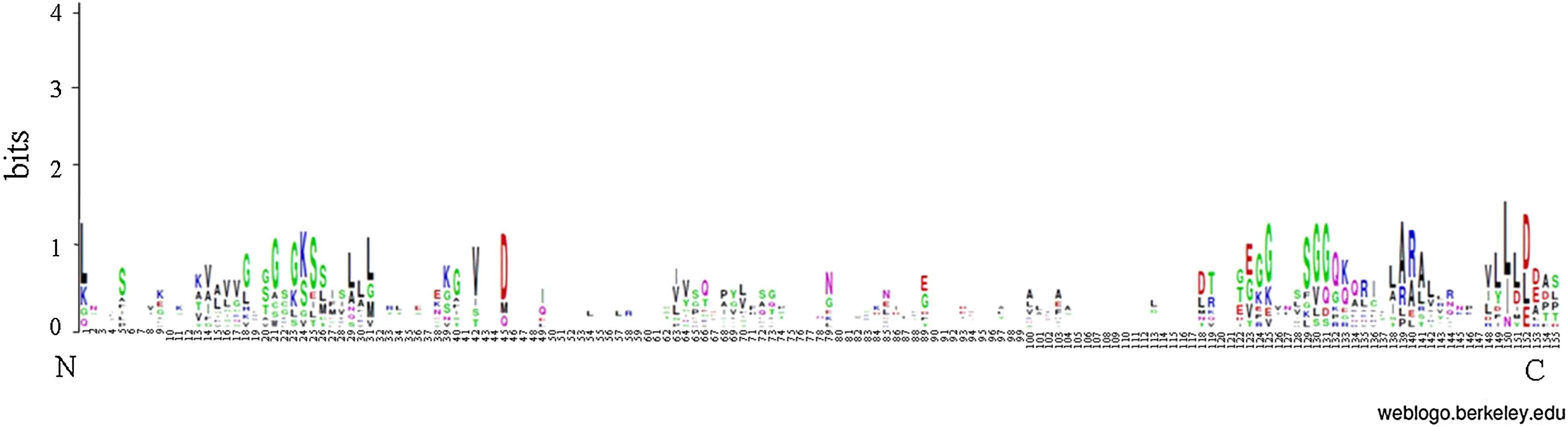
Fig.1 DNA binding domain recognition marker
As we expected, proteins in this family, especially those that are very similar in evolutionary relationships, have identical or similar conserved domains. Based on the analysis of these sequences on the Pfam website, we obtained two conserved protein sequence motifs. The results of the annotation indicate that motif 1 has an ABC transporter transmembrane region. Motif 2 functions as an ABC transporter (Table S4). Based on the analysis of the Pfam website, all proteins of the Bx- mrp gene family contain motif 2. Furthermore, except for Bx- mrp8, the remaining protein sequences in this family all contain motif 1 (Fig.S3).
Hydrophobicity of proteins
The results of the protein hydrophobicity analysis are shown in Fig.2, and the following description takes Bx- mrp1 asan example. The 865th lysine (K) in the Bx- mrp1 protein has a low hydrophobicity score of ? 3.622; thus, it is highly hydrophilic. In contrast, the leucine (L) at position 418 has a higher hydrophobicity score of 3.789, indicating that it is more hydrophobic. The results of the hydrophobic analysis of the remaining nine amino acids in this family are listed in Table S5. From the results of the hydrophobic analysis, the 10 proteins contained numerous hydrophilic and hydrophobic regions, but the distribution and aggregation were not significant.
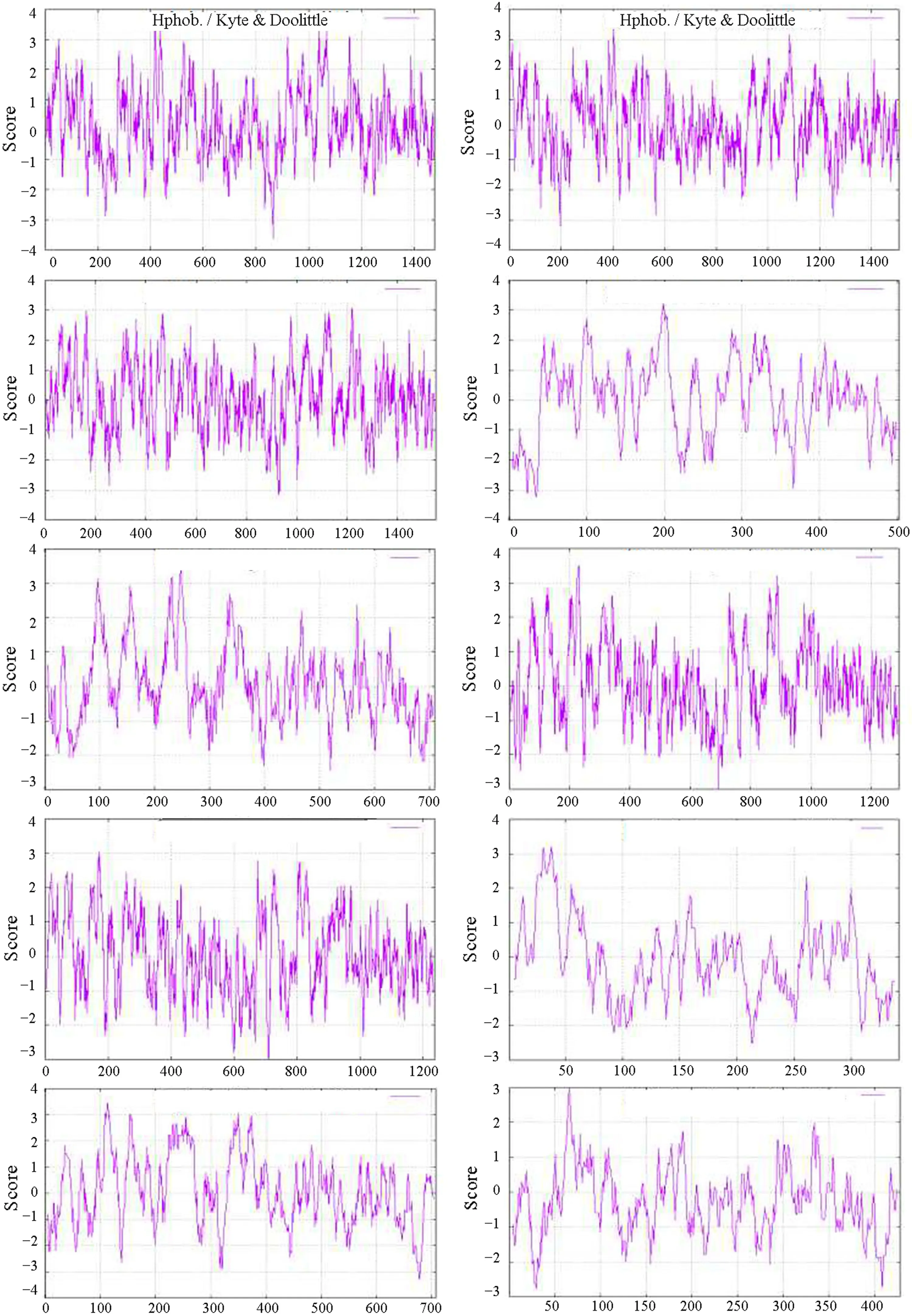
Fig.2 Hydrophobicity of protein
Transmembrane helical segments analysis
The transmembrane helical segments of the Bx- mrp gene family were predicted using the TMHMM website. The relevant results are shown in Fig.3. According to the analysis results, the Bx- mrp2 protein has the most transmembrane helical segments (14). In contrast, the the Bx- mrp10 protein had the fewest (0). In the remaining eight protein sequences, these segments differed in length and number. For example, in the Bx- mrp1 protein sequence, the start and end positions of its twelve transmembrane helix segments were 38–60, 103–125, 132–154, 424–446, 514–531, 546–568, 917–939, 959–981, 1029–1051, 1055–1077, 1139–1161 and 1166–1183, respectively. The sequence starting and ending positions of the extramembranous portion are 61–102, 155–423, 532–545, 940–958, 1052–1054 and 1162–1165, respectively. In contrast, the sequence start positions of the intramembrane portions are 1–37, 126–131, 447–513, 569–916, 982–1028, 1078–1138 and 1184–1477, respectively. Detailed information on the transmembrane helical segments of the other nine protein sequences is in Table S6.
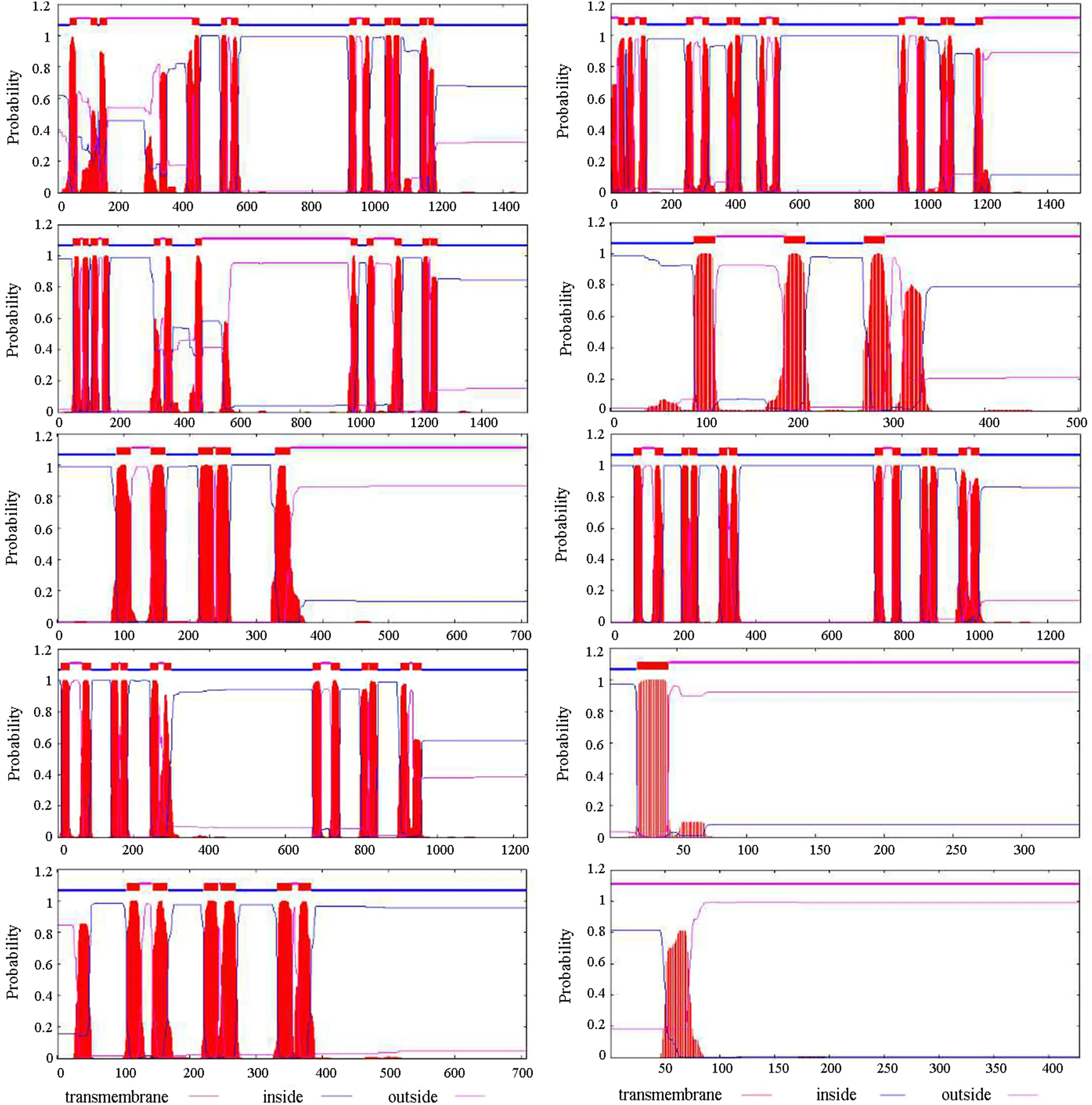
Fig.3 Transmembrane helical segments analysis
Phosphorylation site prediction
The predicted results of phosphorylation sites in this protein family are shown in Fig.4. Again we use Bx- mrp1 to describe a specific prediction results for protein phosphorylation sites. The protein has 89 serine phosphorylation sites (at amino acids 18, 61, 93, 126, 160, 166, 190, 211, 216, 218, 227, 275, 292, 360, 374, 376, 391, 409, 410, 428, 510, 519, 534, 535, 575, 598, 599, 608, 618, 622, 634, 644, 648, 669, 677, 683, 731, 739, 758, 777, 790, 845, 851, 856, 857, 860, 864, 871, 874, 875, 879, 883, 904, 932, 943, 982, 986, 1050, 1052, 1053, 1085, 1086, 1088, 1090, 1105, 1132, 1133, 1136, 1159, 1162, 1178, 1188, 1195, 1203, 1225, 1240, 1245, 1255, 1280, 1281, 1341, 1374, 1377, 1394, 1405, 1425, 1440, 1466 and 1470); 46 threonine phosphorylation sites (at amino acids 26, 120, 214, 238, 243, 266, 316, 363, 382, 399, 450, 532, 541, 550, 611, 613, 689, 721, 767, 788, 853, 858, 886, 898, 929, 948, 1009, 1023, 1045, 1047, 1072, 1077, 1104, 1150, 1176, 1211, 1246, 1275, 1285, 1317, 1329, 1410, 1417, 1436, 1456 and 1474); 7 tyrosine phosphorylation sites (at amino acids 194, 220, 367, 699, 855, 1131 and 1210). Table S7 details the prediction results for the remaining nine proteins in this family.
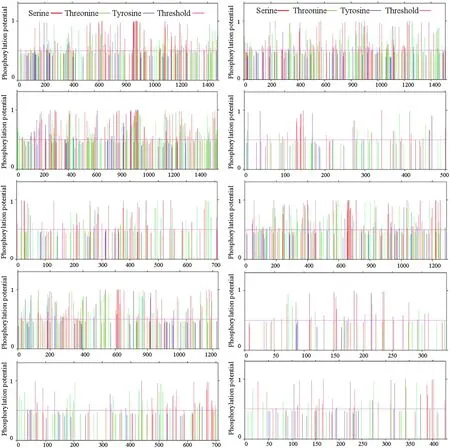
Fig.4 Phosphorylation site prediction
Topological heterogeneity model prediction
The topological prediction results for each gene in this family are shown in Fig.S4. Based on the analysis, Bx- mrp1 and Bx- mrp2 had the most N-glycosylation sites in this family (12); Bx- mrp6 and Bx- mrp10 had the fewest (2). Topological heterogeneity results for other proteins in this family are shown in Table S8.
RT-qPCR verification of the differential gene expression for this family
Based on the RT-qPCR of the 10 genes, Fig.5 shows that the 10 gene members in the mrp family studied here were indeed differentially expressed after treatment with emamectin benzoate, avermectin and matrine. The results of RT-qPCR in Fig.5 show that after treatment with emamectin benzoate, the expression of Bx- mrp5 and Bx- mrp9 and the other eight genes increased, indicating that they are typically resistant to emamectin benzoate. Among them, After treatment with 50 μg mL?1emamectin benzoate, the expression ratio of Bx- mrp6 was the highest, followed by Bx- mrp4 gene, but the expression ratio of Bx- mrp6 was highest after treatment condition of with 100 μg mL?1. However, in response to 150 μg mL?1and 200 μg mL?1, the expression ratio of Bx-
mrp4 was the highest (about 1.53 times and 1.7 times higher, respectively, compared with no exposure). Related results are shown in Fig.S5. From the above results, Bx- mrp4 and Bx- mrp6 genes of the B. xylophilus have strong resistance to emamectin benzoate, and their drug resistance increases with an increase in drug concentration.
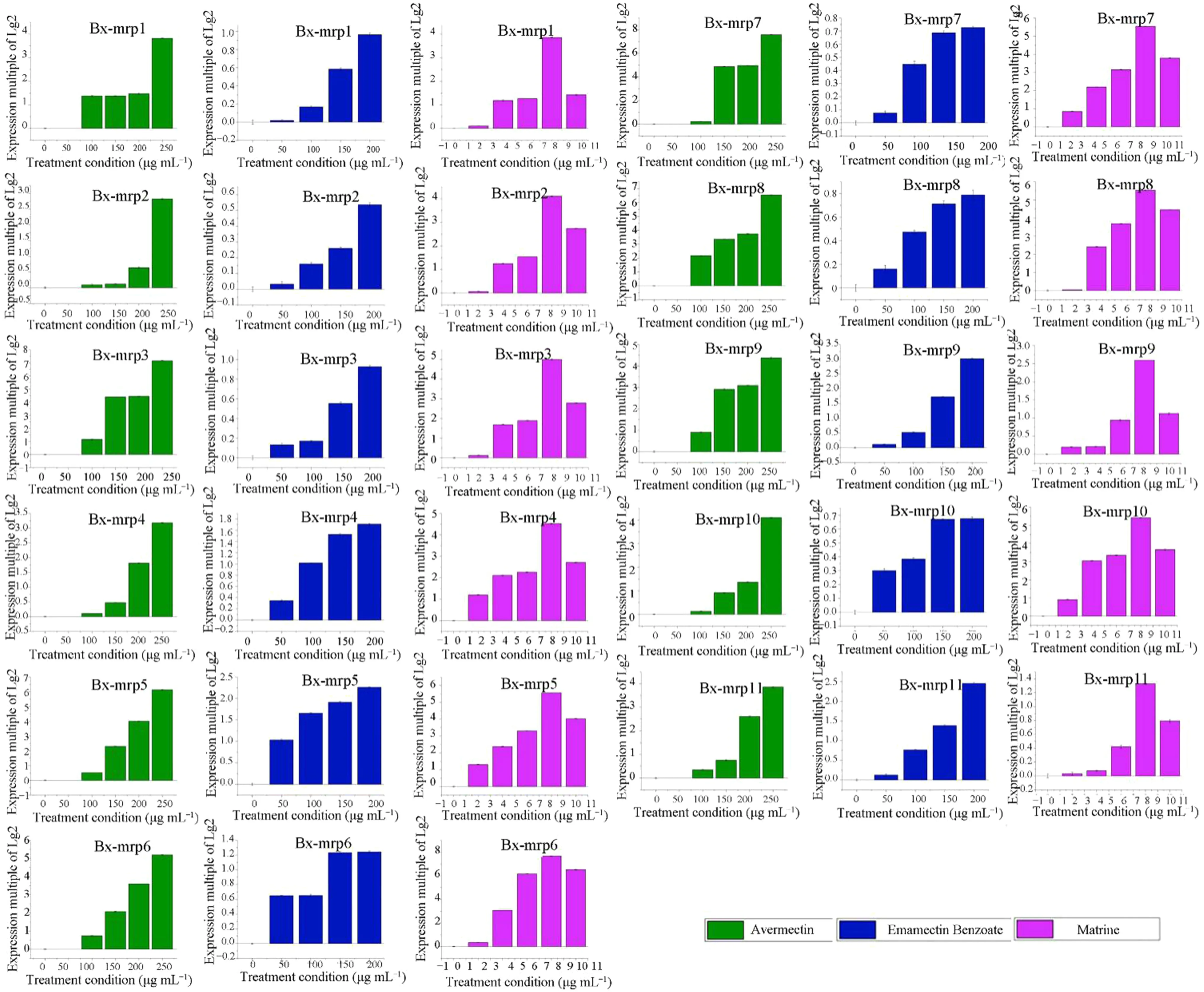
Fig.5 Differential expression of mrp gene family in drug stress
After avermectin treatment, the expression levels of all genes but Bx- mrp9 increased as the concentration of avermectin increased, indicating that they are typically resistant to the avermectin (Fig.5). In response to 100 μg mL?1avermectin, the expression ratio of Bx- mrp8 was the highest, about 2.16 times that with no treatment, whereas the expression ratio of Bx- mrp7 was the highest in response to 150, 200 and 250 μg mL?1(about 4.88, 4.96 and 7.58 times higher than that with no treatment), followed by Bx- mrp3 (about 4.39, 4.44 and 7.16 times higher, respectively, than that with no treatment). Related results are shown in Fig.S5. Thus, Bx- mrp7 and Bx- mrp3 genes have strong avermectin resistance in B. xylophilus, and their drug resistance increases with increasing drug concentrations.
Figure 5 shows that after matrine treatment, the expression levels of nine genes varied consistently; Bx- mrp9 was negatively regulated. With increasing matrine concentrations, the expression of the nine genes first increased, then decreased. Among the six matrine concentrations, the highest expression was found at 8 mg mL?1, indicating that the family members have typical resistance to matrine. After 2 mg mL?1matrine treatment, the expression ratio of Bx- mrp5 gene was the highest, about 1.31 times with no treatment, followed by Bx- mrp4 gene, about 1.2 times that of the untreated control. After treatment with 4, 6 and 8 mg mL?1, the expression ratio of Bx- mrp6 gene was the highest (about 3.07, 6.155 and 7.67 times higher than the control). After the 10 mg mL?1matrine treatment, the expression ratio of Bx- mrp7 gene was the highest, about 7.58 times that of the control. Other relevant results are shown in Fig.S5. From the above results, Bx- mrp6 and Bx- mrp7 genes have strong matrine resistance in B. xylophilus, and the drug resistance increased with an increase in the drug concentration.
Based on the results for all drug treatments, Bx- mrp6 and Bx- mrp7 had strong comprehensive drug resistance after the various drug treatments and gave significant responses to the different drug stresses.
Discussion
The pine wood nematode is concealed in its host, which leads to an extremely complicated relationship with its host (Li et al. 2019a, b). Due to the lack ofeffective control methods, it has caused huge forestry losses. The discovery and utilization of potential control targets is a low-cost and effective strategy to develop effective control measures (Wang et al. 2016). In eukaryotes, the mrp family of genes can remove drugs that enter the cell from another cell, thereby reducing the concentration of the compound in the cell, thus playing a crucial role in the cell’s resistance to a drug. Thus, the role of the mrp gene family in drug resistance and metabolism has attracted increasing attention (Gameiro et al. 2017). The drug-resistant metabolic reaction process of plant parasitic nematodes is their main way to resist drugs. Therefore, the various functions of the mrp gene family in the pine wood nematode need to be studied to provide a theoretical basis for the development of new control technologies.
Toward developing this basic foundation of knowledge, we thus analyzed members of mrp gene family in the pine wood nematode and found that the encoded proteins contain multiple transmembrane domains (TMD) and nucleotidebinding domains (NBD), suggesting that the family of proteins may be a transmembrane protein that can transport substances and function in combination with macromolecular compounds (Dermauw and Van Leeuwen 2014). Our motif analysis revealed that members of the Bx- mrp gene family encode the ABC transporter and the ABC transporter transmembrane region, which can bind extracellular ligands.
In our verification of the function of the Bx- mrp gene family members of the pine wood nematode using RT-qPCR, the expression ofeach gene in the family increased gradually with increasing concentrations of acevar and avermectin and first increased and then decreased with increasing concentrations of matrine (with an overall increasing trend). These results further confirmed our hypothesis that the gene family has typical resistance to drug resistance, but the specific mechanisms involved in the multidrug resistance needs further elucidation.
There have been few comparisons of drug resistance among mrp gene family members in the pine wood nematode (Hao et al. 2018). As mentioned earlier, only Bx-mrp1 has been reported on resistance studies (Hao et al. 2018). Our study showed that members of the pine wood nematode mrp gene family have significant resistance properties. These genes can be potential targets for controlling this destructive plant parasitic nematode. Although our results provide theoretical guidance for the control of pine wood nematode, further exploration is needed to discover other unknown genes and pathways associated with B. xylophilus resistance to eventually develop a biopharmaceutical synergist using Bx-mrp family protein as a biological control target.
References
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:W39–W49
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Dermauw W, Van Leeuwen T (2014) The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochem Mol 45:89–110
Domenichini A, Adamska A, Falasca M (2019) ABC transporters as cancer drivers: potential functions in cancer development. Biochim Biophys Acta General Subjects 1863:52–60
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285
Futai K (2013) Pine Wood Nematode, Bursaphelenchus xylophilus. Annu Rev Phytopathol 51:61–83
Gameiro M, Silva R, Rocha-Pereira C, Carmo H, Carvalho F, Bastos MDL, Remiao F (2017) Cellular models and in vitro assays for the screening of modulators of p-gp, MRP1 and BCRP. Molecules 22(4):600
Hao X, Wang F, Ma L, Wang BW, Ma Y (2018) Cloning and function of resistance gene of Bursaphelenchus xylophilus. J. North East For. Univ. 46(89–92):97
He ZL, Zhang HK, Gao SH, Lercher MJ, Chen WH, Hu SN (2016) Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res 44:W236–W241
Hyde SC, Emsley P, Hartshorn MJ, Mimmack MM, Gileadi U, Pearce SR, Gallagher MP, Gill DR, Hubbard RE, Higgins CF (1990) Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346:362–365
Kikuchi T, Cotton JA, Dalzell JJ, Hasegawa K, Kanzaki N, McVeigh P, Takanashi T, Tsai IJ, Assefa SA, Cock PJA, Otto TD, Hunt M, Reid AJ, Sanchez-Flores A, Tsuchihara K, Yokoi T, Larsson MC, Miwa J, Maule AG, Sahashi N, Jones JT, Berriman M (2011) Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog 7(9):e1002219
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kwon T (2008) Change of abundance of arthropods in pine forests caused by aerial insecticide spray. Arch Environ Contam Toxicol 54:92–106
Li YX, Meng FL, Deng X, Wang X, Feng YQ, Zhang W, Pan L, Zhang XY (2019a) Comparative transcriptome analysis of the pinewood nematode Bursaphelenchus xylophilus reveals the molecular mechanism underlying its defense response to host-derivedpinene. Int J Mol Sci 20(4):911
Li YL, Zheng CY, Liu KC, Wu Y, Fan B, Han ZM (2019b) Transformation of multi-antibiotic resistant Stenotrophomonas maltophilia with GFP Gene to enable tracking its survival on pine trees. Forests 10(3):231
Liu HB, Wu F, Wu XQ, Ye JR (2019a) Differential effects of rapamycin on Bursaphelenchus xylophilus with different virulence and differential expression of autophagy genes under stresses in nematodes. Acta Biochim Biophys Sin 51:254–262
Liu MJ, Hwang B, Jin CZ, Li WJ, Park DJ, Seo S, Kim CJ (2019b) Screening, isolation and evaluation of a nematicidal compound from actinomycetes against the pine wood nematode, Bursaphelenchus xylophilus. Pest Manag Sci 75:1585–1593
Ma YF, Wu ZH, Gao M, Loor JJ (2018) Nuclear factor erythroid 2-related factor 2-antioxidant activation through the action of ataxia telangiectasia-mutated serine/threonine kinase is essential to counteract oxidative stress in bovine mammary epithelial cells. J Dairy Sci 101:5317–5328
Mota MM, Vieira P (2008) Pine wilt disease: a worldwide threat to forest ecosystems. Springer, Berlin
Omasits U, Ahrens CH, Mueller S, Wollscheid B (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886
Seddigh S (2017) Comprehensive comparison of two protein family of P-ATPases (13A1 and 13A3) in insects. Comput Biol Chem 68:266–281
Seddigh S, Darabi M (2018) Functional, structural, and phylogenetic analysis of mitochondrial cytochrome b (cytb) in insects. Mitochondrial DNA A 29:236–249
Soliman T, Mourits MCM, van der Werf W, Hengeveld GM, Robinet C, Lansink AGJM (2012) Framework for modelling economic impacts of invasive species, applied to pine wood nematode in Europe. PLOS ONE 7(9):e45505
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang XR, Cheng X, Li YD, Zhang JA, Zhang ZF, Wu HR (2012) Cloning arginine kinase gene and its RNAi in Bursaphelenchus xylophilus causing pine wilt disease. Eur J Plant Pathol 134(3):521–532
Wang F, Li DL, Ma L, Wang BW, Chen QL, Zhang RZ, Su D, Kang XY, Zhai W (2016) Identification and RNAi of a Bursaphelenchus xylophilus dauer formation gene: Bx-DAF6. For Stud China 38:21–27
Wang BW, Ma L, Wang F, Wang BY, Hao X, Xu JY, Ma Y (2017) Low temperature extends the lifespan of Bursaphelenchus xylophilus throuth the cGMP pathway. Int J Mol Sci 18(11):2320
Wang BW, Hao X, Xu JY, Ma Y, Ma L (2019a) Transcriptome-Based analysis reveals a crucial role of BxGPCR17454 in low temperature response of pine wood nematode ( Bursaphelenchus xylophilus). Int J Mol Sci 20(12):2898
Wang BY, Wen RR, Ma L, Ma Y, Zhou TH (2019b) Molecular characterization and functional analysis of daf-16-2b gene in the pine wood nematode, Bursaphelenchus xylophilus. For Pathol 49(2):e12467
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552
Yang XG, Jia LY, Wei L, Zuo WS, Song SP (2002) Correlation of expression levels of multidrug resistance gene 1 ( mdr 1) mRNA, multidrug resistance-associated protein (MRP), amd P-glycoprotein (P-gp) with chemotherapy effi cacy in malignant lymphomas. Zhonghua Yi Xue Za Zhi 82:1177–1179
Zhang HK, Gao SH, Lercher MJ, Hu SN, Chen WH (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40:W569–W572
Zhao BG (2008) Pine wilt disease in China. In: Zhao BG, Futai K, Sutherland JR, Takeuchi Y (eds) Pine wilt disease. Springer, Tokyo
 Journal of Forestry Research2021年2期
Journal of Forestry Research2021年2期
- Journal of Forestry Research的其它文章
- Stem taper functions for Betula platyphylla in the Daxing’an Mountains, northeast China
- Leaf phenotypic variation of endangered plant Tetracentron sinense Oliv. and influence of geographical and climatic factors
- Ecological variations of woody species along an altitudinal gradient in the Qinling Mountains of Central China: area-based versus mass-based expression of leaf traits
- Variations in stem radii of Larix principis-rupprechtii to environmental factors at two slope locations in the Liupan Mountains, northwest China
- A review of ecological mechanisms for management practices of protective forests
- Variation of basic density, calorific value and volumetric shrinkage within tree height and tree age of Ugandan grown Eucalyptus grandis wood
