Na2CO3-responsive Photosynthetic and ROS Scavenging Mechanisms in Chloroplasts of Alkaligrass Revealed by Phosphoproteomics
Jinwei Suo,Heng Zhang,Qi Zhao,Nan Zhang,Yongxue Zhang,Ying Li,Baohua Song,Juanjuan Yu,Jianguo Cao,Tai Wang,Ji Luo,Lihai Guo,Jun Ma,Xumin Zhang,Yimin She,Lianwei Peng,Weimin Ma,Siyi Guo,Yuchen Miao,Sixue Chen,9,Zhi Qin,,Shaojun Dai,,
1 Development Center of Plant Germplasm Resources,College of Life and Environmental Sciences,Shanghai Normal University,Shanghai 200234,China
2 Alkali Soil Natural Environmental Science Center,Northeast Forestry University,Key Laboratory of Saline-alkali Vegetation Ecology Restoration in Oil Field,Ministry of Education,Harbin 150040,China
3 State Key Laboratory of Subtropical Silviculture,Zhejiang A &F University,Hangzhou 311300,China
4 Institute of Botany,Chinese Academy of Sciences,Beijing 100093,China
5 AB Sciex Asia Pacific Application Support Center,Shanghai 200233,China
6 Shanghai Center for Plant Stress Biology,Chinese Academy of Sciences,Shanghai 201602,China
7 School of Life Sciences,Fudan University,Shanghai 200433,China
8 Institute of Plant Stress Biology,State Key Laboratory of Cotton Biology,Department of Biology,Henan University,Kaifeng 475004,China
9 Department of Biology,Genetics Institute,Plant Molecular and Cellular Biology Program,Interdisciplinary Center for Biotechnology Research,University of Florida,Gainesville,FL 32610,USA
Abstract Alkali-salinity exerts severe osmotic,ionic,and high-pH stresses to plants.To understand the alkali-salinity responsive mechanisms underlying photosynthetic modulation and reactive oxygen species (ROS) homeostasis,physiological and diverse quantitative proteomics analyses of alkaligrass (Puccinellia tenuiflora)under Na2CO3 stress were conducted.In addition,Western blot,real-time PCR,and transgenic techniques were applied to validate the proteomic results and test the functions of the Na2CO3-responsive proteins.A total of 104 and 102 Na2CO3-responsive proteins were identified in leaves and chloroplasts,respectively.In addition,84 Na2CO3-responsive phosphoproteins were identified,including 56 new phosphorylation sites in 56 phosphoproteins from chloroplasts,which are crucial for the regulation of photosynthesis,ion transport,signal transduction,and energy homeostasis.A full-length PtFBA encoding an alkaligrass chloroplastic fructosebisphosphate aldolase (FBA) was overexpressed in wild-type cells of cyanobacterium Synechocystis sp.Strain PCC 6803,leading to enhanced Na2CO3tolerance.All these results indicate that thermal dissipation,state transition,cyclic electron transport,photorespiration,repair of photosystem (PS) II,PSI activity,and ROS homeostasis were altered in response to Na2CO3stress,which help to improve our understanding of the Na2CO3-responsive mechanisms in halophytes.
KEYWORDS Chloroplasts;Na2CO3stress;ROS scavenging;Phosphoproteomics;Puccinellia tenuiflora
Introduction
Soil salinization and alkalization frequently occur simultaneously.In northeast China,more than 70% of the land area has become alkaline grassland [1].Alkali-salinity is one of the most severe abiotic stresses,limiting the productivity and geographical distribution of plants.Saline-alkali exerts osmotic stress and ion damage,as well as high-pH stress to plants[2].However,little attention has been given to the sophisticated tolerance mechanisms underlying plant response to saline-alkali (e.g.,Na2CO3and NaHCO3) stresses [3,4].As the organelle for photosynthesis,chloroplasts are extremely susceptible to saline-alkali stress [5].Excessive accumulation of Na+reduces the CO2diffusion through stomata and mesophyll,negatively affecting plant photosynthesis[6].As a consequence,excessive excitation energy causes generation of reactive oxygen species(ROS),resulting in damage to the thylakoid membrane [6].
Current high-throughput proteomic approaches are powerful to untangle the complicated mechanisms of chloroplast development,metabolism,and stress response [7-10].More than 522 NaCl-responsive chloroplast proteins were found in different plant species,such as tomato(Solanum lycopersicum)[11],wheat(Triticum aestivum)[12],and other plant species[13-18].The presence of these proteins indicate that the light harvesting,photosynthetic electron transfer,carbon assimilation,ROS homeostasis,energy metabolism,signaling,and membrane trafficking were modulated in chloroplasts in response to NaCl stress.However,only about 53 salinity-responsive genes encoding chloroplast proteins have been characterized[5],which are insufficient to address the sophisticated salinityresponsive networks in chloroplasts.Additionally,NaCl stress altered phosphorylation levels of several chloroplast proteins in Arabidopsis [19,20],Brachypodium distachyon[21],and sugar beet (Beta vulgaris) [22],implying that state transition,PSII damage repair,thermal dissipation,and thylakoid membrane organization were crucial for plant acclimation to salt stress[23].However,the critical roles of reversible protein phosphorylation in salinity-/alkali-responsive metabolic networks are virtually unknown.
Alkaligrass (Puccinellia tenuiflora) is a monocotyledonous halophyte species belonging to the Gramineae,and is widely distributed in the Songnen Plain in Northeastern China.It has strong ability to survive in extreme saline-alkali soil (pH range of 9-10).Several salinity-/alkali-responsive genes and/or proteins in leaves and roots of alkaligrass have been reported [24-26].A previous transcriptomic study also revealed that a number of Na2CO3responsive genes were overrepresented in metabolism,signal transduction,transcription,and cell rescue [27].Despite this progress,the precise alkali-responsive mechanisms in chloroplasts are still poorly understood.Analyses of the photosynthetic and ROS scavenging mechanisms in chloroplasts regulated by the reversible protein phosphorylation and the expression of nuclear and chloroplast genes are critical for understanding the Na2CO3-responsive mechanisms in alkaligrass.In this study,we investigated the alkali-responsive characteristics in chloroplasts and leaves of alkaligrass.By integrative analyses of protein phosphorylation,patterns of protein abundance,gene expression,photosynthesis parameters,antioxidant enzyme activities,and chloroplast ultrastructure,we revealed several important Na2CO3-responsive strategies in the halophyte alkaligrass.These results have yielded important insights into the alkaliresponsive mechanisms in halophytes.
Results
Na2CO3treatment decreased seedling growth and biomass
Na2CO3treatment clearly affected the morphology and biomass of alkaligrass seedlings.The leaves withered with the increase of Na2CO3concentration and treatment time(Figure S1).The shoot length and relative water content decreased significantly at 200 mM Na2CO3of 24 h after treatment (24 HAT200) (Figure 1A).The fresh and dry weights of leaves also clearly decreased under 200 mM Na2CO3treatment(Figure 1B).
Na2CO3treatment changed ionic and osmotic homeostasis,cell membrane integrity,and abscisic acid level
Na2CO3treatment perturbed the ion and pH homeostasis in leaves.Na+in leaves was gradually accumulated,but K+content did not show obvious changes,resulting in the sharp decline of the K+/Na+ratio (Figure 1C,D).In addition,the Mg2+and Ca2+contents gradually decreased under the Na2CO3treatment (Figure 1E).Malondialdehyde content and relative electrolyte leakage significantly increased under different Na2CO3treatments,indicating that the membrane integrity was affected by Na2CO3treatment (Figure 1F).In addition,proline and glycine betaine gradually accumulated with the increase of Na2CO3concentrations(Figure 1G),while the soluble sugar content only showed marked accumulation at 200 mM Na2CO3(Figure 1H).The endogenous abscisic acid(ABA) content in leaves increased significantly (Figure 1H).
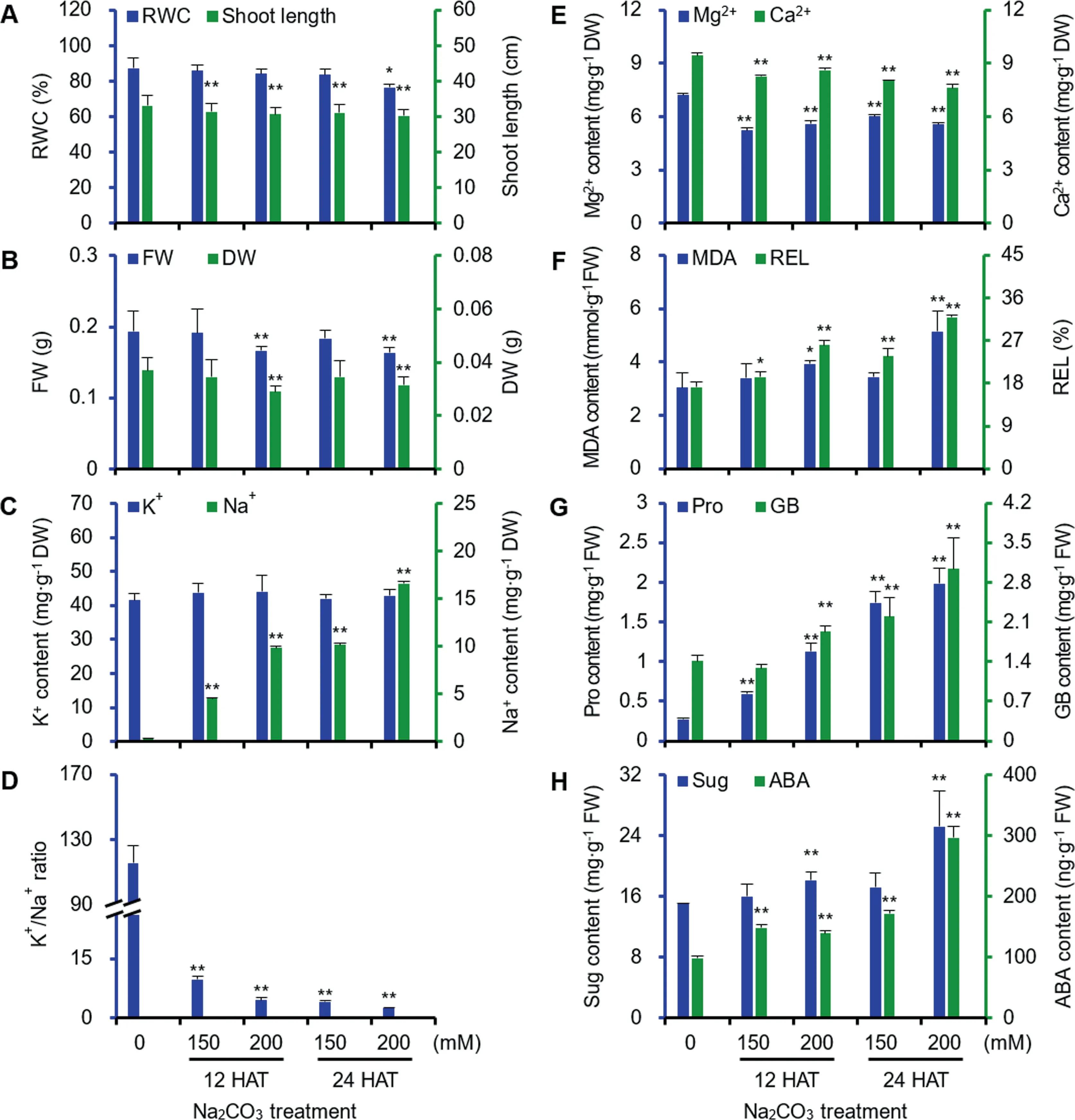
Figure 1 Leaf physiological characteristics in alkaligrass under Na2CO3treatment
Photosynthesis and chlorophyll content decreased under Na2CO3
In seedlings,net photosynthetic rate,stomatal conductance,and transpiration rate (Figure 2A and B) gradually decreased,while the intercellular CO2concentration did not exhibit obvious changes under the Na2CO3treatment (Figure 2B).
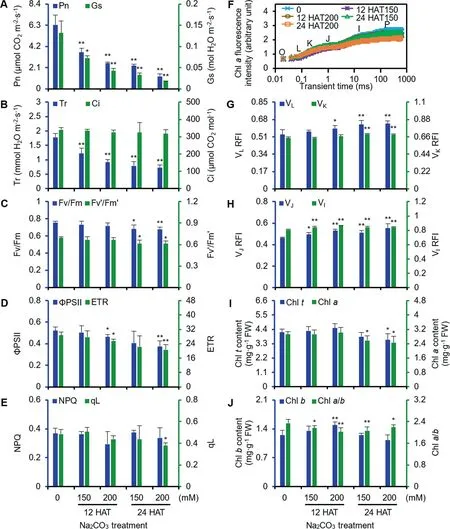
Figure 2 Photosynthetic characteristics of alkaligrass under Na2CO3treatment
To evaluate the photosynthetic performance,we investigated the changes of chlorophyll (Chl) fluorescence and the polyphasic fluorescence transients (OJIP).The maximum quantum efficiency of PSII photochemistry and PSII maximum efficiency significantly decreased at 24 HAT(Figure 2C),and the actual PSII efficiency and electron transport rate were declined remarkably at 200 mM Na2CO3(Figure 2D).In addition,the non-photochemical quenching did not change and the fraction of open PSII centers significantly decreased at 24 HAT200 (Figure 2E).The fluorescence transient gradually decreased,reaching the lowest level at 24 HAT200 (Figure 2F).After normalization,the relative fluorescence intensities of VLand VK,two specific indicators of thylakoid dissociation and oxygen-evolving complex (OEC) damage increased at 24 HAT (Figure 2G).VJand VI,however,significantly increased.The relative variable fluorescence intensity of VJand VIcan be considered as a measurement of the accumulation of QA-and the proportion of the QB-nonreducing reaction center.This suggests that the accumulation of QA-and increased proportion of QB-non-reducing reaction center in the Na2CO3-stressed leaves (Figure 2H).In addition,the contents of total Chl and Chladecreased at 24 HAT (Figure 2I),and the ratio of Chla/balso decreased under the Na2CO3treatment (Figure 2J).
Na2CO3treatment affected chloroplast ultrastructure
Na2CO3treatment changed the chloroplast ultrastructure in mesophyll cells and bundle sheath cells from lateral veins,minor veins,and midveins (Figure3).Under normal conditions,chloroplasts in mesophyll cells and bundle sheath cells exhibited long ellipsoidal or shuttle-shaped,double membrane compartment,with only a few osmophilia plastoglobules in the stroma (Figure 3A,F,K,and P).Thylakoifirst-strand cDNA was obtainedds were dispersed in the chloroplasts,and the fully developed thylakoid membrane systems were well organized in grana and stromal lamellae (Figure 3A,F,K,and P).At 12 HAT,slight swelling of the chloroplast stroma occurred,and the membranes of the individual thylakoid fused,eliminating the intraspace (Figure 3C,H,and M).While at 24 HAT,chloroplast volume increased obviously to become round-shaped.The thylakoid membrane systems in various cells became distorted and incomplete,showing a dilated intraspace (Figure 3D,E,J,and O).The size and number of grana somewhat decreased,and some grana completely disappeared (Figure 3D,E,and J).At 24 HAT200,numerous plastoglobules were observed in chloroplasts,and the size and number of plastoglobules appeared to be Na2CO3concentration-dependent (Figure 3).This implies that lipid peroxidation-mediated destruction of the thylakoid membranes takes place in chloroplasts.In addition,the aforementioned changes in thylakoids appeared more drastic in mesophyll cells than in the bundle sheath cells (Figure 3D,E).
Na2CO3treatment changed antioxidant enzymes in leaves and isolated chloroplasts
To evaluate the level of oxidative stress in leaves and chloroplasts under Na2CO3treatment,the O2-generation rate,H2O2content and four metabolites (i.e.,ascorbate (AsA),dehydroascorbate (DHA),glutathione (GSH),and oxidized glutathione (GSSG)),and the activities of nine antioxidant enzymes in ROS scavenging system were monitored (Figure4).
In leaves,the O2-generation rate and H2O2content increased under the Na2CO3treatment (Figure 4A).The contents of several metabolites (e.g.,reduced AsA,DHA,and GSSG) did not change,but GSH increased at 24 HAT(Figure 4B,C).Importantly,the activities of superoxide dismutase (SOD),catalase (CAT),and dehydroascorbate reductase (DHAR) decreased at 24 HAT,and CAT activity was inhibited at 12 HAT200 (Figure 4D,E,and F).The activities of peroxidase (POD),ascorbate peroxidase (APX),monodehydroascorbate reductase (MDHAR),glutathione peroxidase (GPX),glutathione reductase (GR),and glutathione Stransferase (GST) showed increased patterns under the Na2CO3treatment (Figure 4D,E,F,G,and H).These results indicate that the superoxide dismutation by SOD and reduction of H2O2to H2O by CAT decreased,but the APX/POD pathway,AsA-GSH cycle,and GPX pathway were enhanced to cope with the Na2CO3-induced oxidative stress.
We isolated chloroplasts with high purity for activity and proteomics analyses (Figure S2A,B).In isolated chloroplasts,the AsA content decreased,but the DHA content increased under Na2CO3.The contents of GSH and GSSG stayed at relative stable levels at 24 HAT (Figure 4I,J).The activities of SOD,POD,APX,and MDHAR increased at 24 HAT,and APX activity increased at 12 HAT (Figure 4K,L,and M).The activities of DHAR,GPX,and GR were inhibited,however GST activity did not change significantly under Na2CO3(Figure 4L,M,and N).These results indicate that ROS in chloroplasts were mainly dismutated by SOD,and subsequently reduced in APX/POD pathway under the Na2CO3treatment.
Na2CO3-responsive proteome revealed modulation of photosynthesis and ROS scavenging to cope with the stress
A total of 104 Na2CO3-responsive proteins in leaves were identified and classified into ten functional categories (Figure5A,Figure S3,Tables S1-S3).Cluster analysis generated two main clusters (Figure 5B).In Cluster I,63 Na2CO3-decreased proteins were involved in photosynthesis,carbohydrate and energy metabolism,protein synthesis and turnover,and cell wall metabolism.In Cluster II,41 Na2CO3-increased proteins were related to energy metabolism,Chl metabolism,membrane and transport,and cell cycle.Importantly,subcellular localization prediction suggested that 63 proteins (60.6%) were specially localized in chloroplasts,and six proteins (6%) in either chloroplasts or other subcellular locations (Figure 5C;Tables S3 and S4).The changes of 37 photosynthesis-related proteins indicate that the balance of excitation energy between PSII and PSI was disrupted and the efficiency of electron transfer and CO2assimilation were inhibited.In contrast,photorespiration was induced under the Na2CO3treatment.Aside from this,changes of 11 proteins involved in ROS and ion homeostasis as well as signaling pathway were triggered under Na2CO3treatment (Table S3).
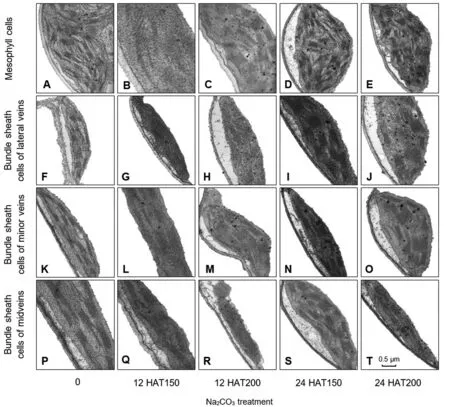
Figure 3 Ultrastructure of chloroplasts in alkaligrass leaves under Na2CO3treatment
Furthermore,we identified 121 Na2CO3-responsive proteins in chloroplasts (Table S5).Among them,there were 102 chloroplast-localized proteins belonging to eight functional categories (Figure 5D;Tables S6 and S7).Of these,49 were photosynthetic proteins accounting for 48% of the total.This included five chlorophylla/bbinding proteins,14 PSIIrelated proteins,seven PSI-related proteins,nine photosynthetic electron transfer chain proteins,four subunits of ATP synthase and ten Calvin cycle enzymes.Most of them were obviously altered at 12 HAT200 and 24 HAT (Table S6).Besides,nine photosynthetic electron transfer chain proteins and four subunits of chloroplast ATP synthase were changed(Table S6).This indicates that although Na2CO3inhibited the light harvesting,the PSII and PSI were not changed much at 12 HAT,but were enhanced at 24 HAT.However,ATP synthesis decreased at 12 HAT200,and then recovered to normal or enhanced at 24 HAT.Changes of the ten Calvin cyclerelated proteins imply that carbon assimilation was inhibited at 24 HAT (Table S6).In addition,among the five ROS scavenging enzymes,thioredoxin peroxidase,2-Cys peroxiredoxin BAS1,and GR increased at 12 HAT and decreased at 24 HAT,while APX and GST decreased at 12 HAT and not changed at 24 HAT (Table S6).
Phosphoproteomics revealed novel Na2CO3-responsive phosphorylation sites
We identified 63 Na2CO3-responsive phosphoproteins in leaves.Of these,39 proteins showed increased phosphorylation levels and 21 had decreased phosphorylation levels.These proteins were classified into seven functional categories (Figure 5E,Table S8).Thirty-four phosphoproteins were predicted to be chloroplast-located,and involved in light harvesting,PSII,Calvin cycle and ATP synthesis (Table S8).
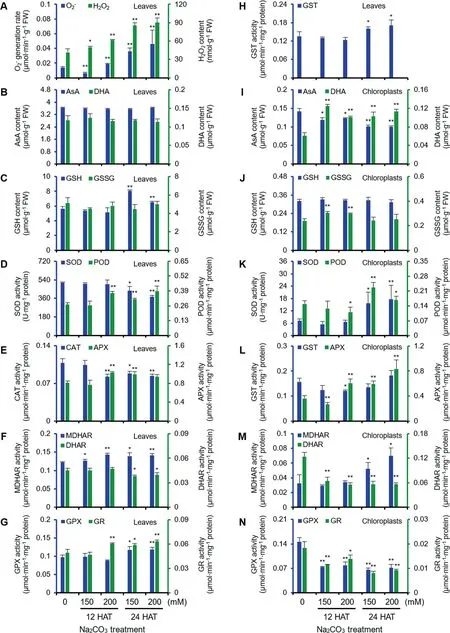
Figure 4 Effects of Na2CO3treatments on antioxidant systems in leaves and chloroplasts of alkaligrass
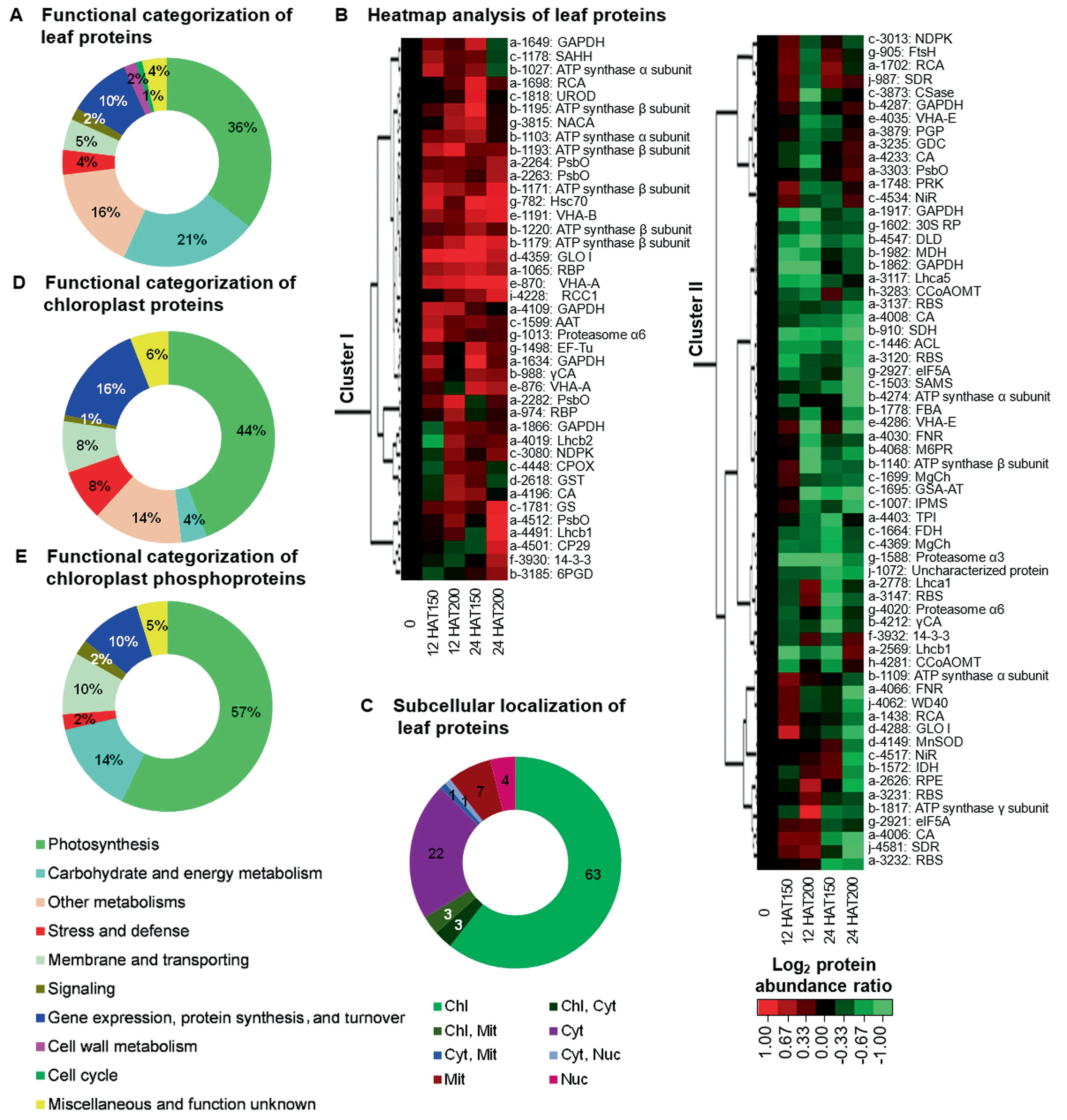
Figure 5 Functional categorization,hierarchical clustering analysis,and subcellular location prediction of the Na2CO3-responsive proteins
In chloroplasts from the Na2CO3-treated leaves,161 unique phosphopeptides were identified,and 137 were quantified by dimethyl labeling (Figure 6A and Table S9).Among them,50 proteins were found to be Na2CO3-responsive with 57 phosphorylation sites,including 33 increased and 15 decreased(Figure 6B and Table S10).The increased phosphoproteins include seven light harvesting proteins,six PSII proteins,five PSI proteins,three electron transfer chain proteins,two Calvin cycle-related proteins,a Na+/H+antiporter,a villin-2,and two thylakoid organization related proteins.The decreased include five ATP synthase subunits and sucrosephosphate synthase.In addition,two signaling related proteins and six proteins involved in gene expression and protein turnover increased in phosphorylation at 24 HAT (Table S10).
In summary,we identified a total of 84 Na2CO3-responsive,chloroplast-localized phosphoproteins in leaf and chloroplast phosphoproteomes (Figure 6C,S4,and S5;Table S10).We identified 56 novel phosphorylation sites in the Na2CO3-responsive chloroplastic phosphoproteins,which may be crucial for regulating photosynthesis,membrane and transport,signaling,stress response,and protein synthesis and turnover (Table S11).
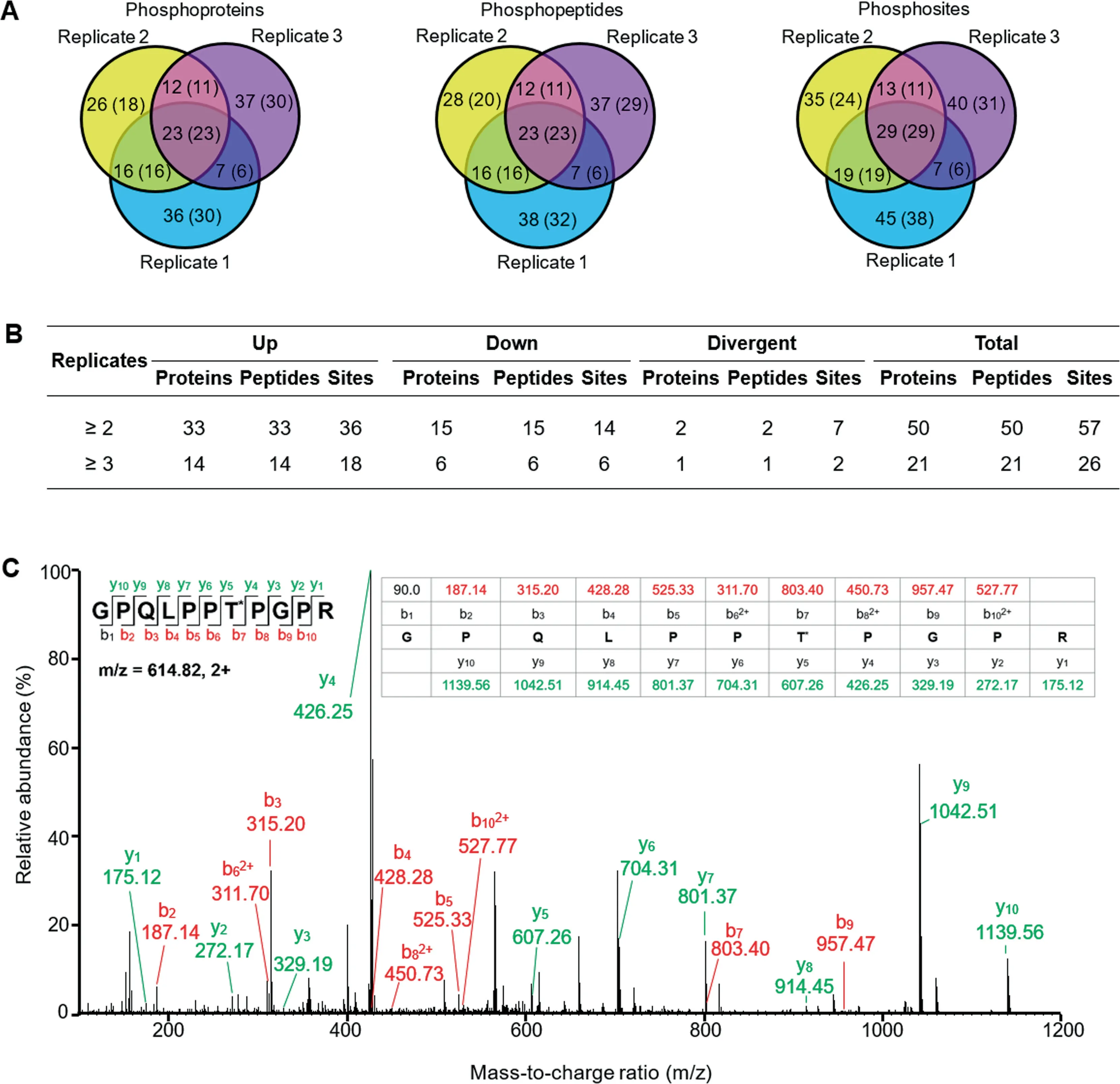
Figure 6 Summary of alkaligrass chloroplast phosphoproteome
Three-dimensional (3D) structure modeling of phosphoproteins
We built thirteen homology-based 3D models of chloroplastlocalized phosphoproteins using the SWISS-MODEL program (Figure S6A,B,C,D,F,G,I,J,K,L,M,and N).We also accepted two experimentally solved 3D structures as homology models by the significant amino acid sequence similarity and conserved phosphorylation sites with our phosphoproteins (Figure S6E,H).The 3D models showed the numbers of helices and beta sheets,and the phosphorylation sites of each protein (Figure S6 and Table S12).
Twenty-eight homologous genes of Na2CO3-responsive phosphoproteins exhibited diverse expression patterns
In order to evaluate the gene expression patterns of the Na2CO3-responsive phosphoproteins,28 homologous genes were analyzed through quantitative real-time PCR (qRTPCR) analysis with ubiquitin as an internal control (Figure S7,Table S13).Ten down-regulated genes were involved in light harvesting,PSII and PSI assembling,photosynthetic electron transfer,ATP synthesis,and thylakoid organization (Figure S7).Besides,three genes (i.e.,photosystemIreactioncentersubunitII(PsaD),serine/arginine-richsplicingfactor33-like,andNa+/H+antiporter) maintained stable levels under the Na2CO3.Interestingly,15 up-regulated genes were involved in photosynthesis,Na+/H+transport,calcium sensing,gene expression,and protein turnover.
Immunodetection of seven representative Na2CO3-rsponsive proteins
To further evaluate the protein abundances of representative photosynthetic proteins under Na2CO3treatment,Western blotting was conducted using available antibodies.The abundances of PSII subunits (photosystem II 22 kDa protein (PsbS),photosystem II D1 protein (D1),and oxygen evolving enhancer protein (PsbO)) and PSI subunit of PsaD increased at 24 HAT200 (Figure7).RuBisCO large subunit (RBL) decreased at 24 HAT (Figure 7).Photosynthetic electron transfer chain related cytochromef(Cytf) and Calvin cycle related phosphoglycerate kinase (PGK) maintained stable protein abundances at 24 HAT.Calvin cycle related sedoheptulose-1,7-bisphosphatase (SBPase) was used as the loading control (Figure 7).
Over-expression of PtFBA enhanced cell alkali tolerance
In our proteomics results,chloroplast-localized FBA increased significantly at phosphorylation level under Na2CO3treatment.Therefore,FBA was selected as a representative Na2CO3responsive protein for functional analysis.The full length cDNA ofPtFBAwas ligated intoPpsbAIIexpression vector,and then transformed to wild-type (WT) cells of a model cyanobacteriumSynechocystis6803,generating aPtFBAover-expression (OE-PtFBA) strain (Figure8A).As expected,PCR analysis confirmed a complete segregation of theOE-PtFBAstrain (Figure 8B).Transcript analysis ofPtFBAgene demonstrated the presence of gene product in theOE-PtFBAcells (Figure 8C).Western blotting analysis using a generic antibody against FBA also demonstrated that the FBA significantly increased in theOE-PtFBAstrain when compared with WT (Figure 8D).The growth of theOE-PtFBAcells,as deduced from cell density and Chlacontent,was much higher than the WT strain under the treatment of 0.4 M Na2CO3for 4 days,although their growth was similar under normal conditions (Figure 8E,F).Thus,we conclude that overexpression ofPtFBAresulted in enhanced Na2CO3tolerance ofSynechocystis6803.
Discussion
Diverse photoprotection mechanisms to counteract Na2CO3-induced photoinhibition
Photosynthesis modulation is critical for plant stress response.PSII supercomplex was very sensitive to environmental changes [28,29].In Na2CO3-treated alkaligrass,PSII (e.g.,OEC and the reaction center proteins) was oxidatively damaged,resulting in the decreases of photochemical efficiency and electron transport [25,30].Our results indicate that diverse photoprotection mechanisms were employed in alkaligrass to counteract alkali-induced photoinhibition.First,the accumulation of PsbS,chlorophylla/bbinding protein (CP) 24,and CP29,as well as induction ofCP24gene at 24 HAT may contribute to PsbS-protonation-dependent conformation conversion of PSII antenna system,suggesting that PsbS-dependent thermal dissipation was enhanced to minimize the potential for photo-oxidative damage under the Na2CO3treatment (Figure S8A) [31].Consistently,CP24 and CP29 also displayed high abundances in salt-sensitive plants (Arabidopsis,oilseed rape (Brassicanapus),and potato (Solanumtuberosum)) and salt-tolerant plants (Indian mustard (Brassicajuncea),mangrove (Kandeliacandel),wild tomato (Solanumchilense),and sugar beet) under salt stresses [17,32-37].Second,the phosphorylation at Ser186 of CP24,Thr165 and Ser172 of CP26,as well as Ser95 and Thr108 of CP29 were enhanced in alkaligrass at 24 HAT (Table S10),while CP24 became dephosphorylated in NaCl-treatedB.distachyon[21].The reversible phosphorylation of CP24 was supposed to regulate the alternative mode of phosphorylation-independent thermal dissipation and phosphorylation-dependent energy spillover in lycophytes [38].Third,the state transition between PSII and PSI was regulated by protein phosphorylation in the Na2CO3-treated alkaligrass (Figure S8B).A serine/threonine-protein kinase (STN7) and protein phosphatases modulate the reversible phosphorylation of LHCII (i.e.,Lhcb1 and Lhcb2) and thylakoid soluble phosphoprotein of 9 kDa (TSP9) [23].The phosphorylation of Lhcb1 was reported to be salinity-increased inB.distachyon[21].Besides,TSP9 interacts with LHCII and the peripheries of PSII and PSI,facilitating dissociation of LHCII from PSII for regulating photosynthetic state transitions [39].Thus,the Na2CO3-enhanced gene expression ofLhcb1,abundances of Lhcb1,Lhcb2,and STN7,as well as phosphorylation of Lhcb1 and TSP9 may facilitate the state transition in alkaligrass (Figure S8B).Similarly,Lhcb1 and Lhcb2 increased in abundances by salt in salt-tolerant plants (e.g.,K.candel,moss (Physcomitrellapatens)),B.juncea,soybean(Glycinemax),and sugar beet [17,34,35,40,41].Furthermore,Lhcb3 phosphorylation also increased in alkaligrass(Table S10).The rate of state transition was induced in ArabidopsisLhcb3knockout mutant [42].This implies that Lhcb3 is probably involved in the state transition,but the underlying regulatory mechanism needs to be further investigated (Figure S8B).
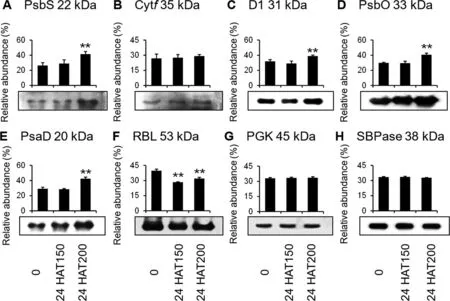
Figure 7 Validation of eight alkali-responsive chloroplast proteins with immunoblotting
Cyclic electron transport (CET) is critical for protecting photosynthetic apparatus and additional ATP supply [43].Several decreased electron transport-related proteins indicate that electron transport was slowed down in alkaligrass at 12 HAT(Figure S8C),and this may alleviate the damage of plastoquinone over-reduction.However,at 24 HAT,when the photosynthetic capacity and electron transport rate were inhibited,CET was induced due to the Na2CO3-increased gene expression ofCyt f,accumulated protein levels and enhanced phosphorylation of ferredoxin-NADP(+) reductase (FNR),Cytfand cytochromeb6fcomplex iron-sulfur subunit(Figure S8C).Among them,FNR phosphorylation may modify its thylakoid membrane localization to regulate the electron transport[44].Additionally,several CET-related proteins(e.g.,Cytf,cytochromeb6fcomplex iron-sulfur subunit,FNR,NAD(P)H-quinone oxidoreductase subunit M (NdhM),and NdhJ) were accumulated in the NaCl-stressed halophytes[14,45].Therefore,the Na2CO3-stressed alkaligrass employed NDH-dependent CET to alleviate photo-oxidative damage and provide extra ATP.
Photorespiration is critical for GSH synthesis,nitrogen and carbon assimilation,and feedback regulation of photosynthetic activity to cope with alkali stress (Figure S8D)[4,25,46].In addition,the decrease of photosynthesis was not resulted from stomatal limitation,but from inhibition of Calvin cycle because most Calvin cycle enzymes decreased significantly (Figure S8D).This is consistent with a previous report in halophytes and salt-tolerant cultivars [4,47].
Enhancement of PSII repair machinery to minimize photodamage
PSII repair machinery was efficiently and dynamically employed to minimize photodamage in alkaligrass(Figure S8E).Na2CO3-induced STN7 and STN8 can promote the phosphorylation of PSII core proteins and CP29.The protein phosphorylation loosens the attractive forces among the subunits of PSII-LHCII supercomplex,enabling migration of the damaged PSII to stroma thylakoids for subsequent detachment of damaged D1 from the core complex during the repairing process [48].Subsequently,the thylakoid lumen 18.3 kDa protein can function as an acid phosphatase to dephosphorylate the damaged D1 protein,and this process is facilitated by the Na2CO3-accumulated PsbO with GTPase activity.The dephosphorylated D1 is recognized and degraded by the Na2CO3-induced ATP-dependent zinc metalloprotease FtsH 2 (FtsH) (Figure S8E).Besides,Na2CO3-induced ToxAbinding protein 1 may contribute to the D1 degradation process through its positive regulatory role in the stabile accumulation of FtsH protease in chloroplast stroma [49].Simultaneously,nascent copies of D1 protein are synthesized and processed rapidly(Figure S8E).Under Na2CO3treatment,the D1 maturation and co-translational insertion into PSII complexes were prompted by the inducedPsbHgene,accumulation of PsbH and low PSII accumulation 1 protein,as well as the induced phosphorylation of Thr3 or Thr5 in the PsbH [50,51].
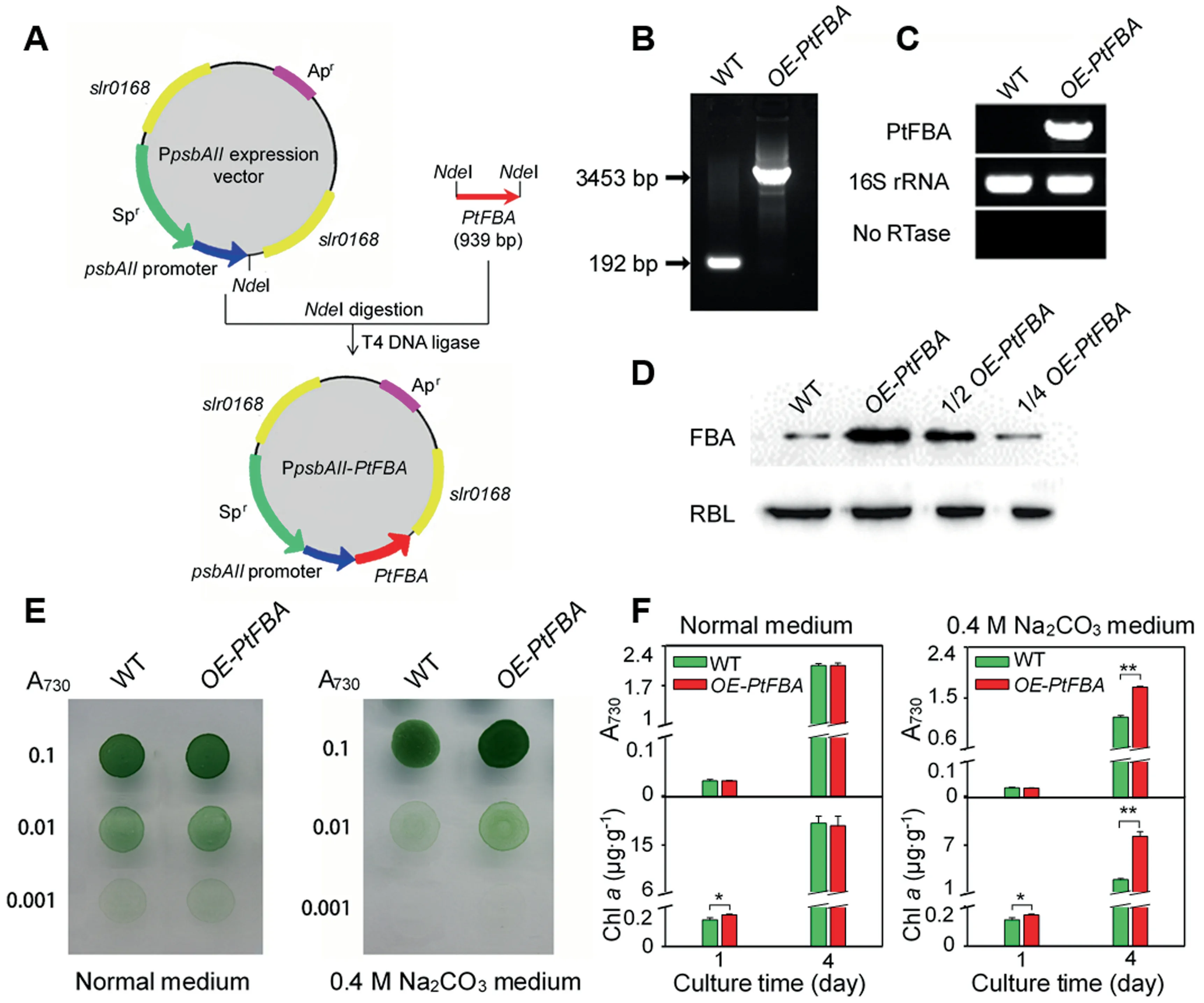
Figure 8 Transgenic analysis of chloroplast-localized PtFBA gene in a model cyanobacterium Synechocystis sp.strain PCC 6803
OEC (PsbO,PsbP,and PsbQ) is peripherally bound to PSII at the luminal side of the thylakoid membrane,which can stabilize the binding of inorganic cofactors,maintain the active Mn-cluster,and enhance oxygen-evolution in PSII [52].The expression ofPsbOandPsbPdecreased in leaves at 24 HAT,and the abundance of OEC was affected in alkaligrass and mangrove [45].Interestingly,phosphorylation of PsbO and PsbP was Na2CO3-decreased in alkaligrass (Figure S8C),and PsbP and PsbQ have been reported to be phosphorylated in thylakoid lumen of Arabidopsis [53].This indicates that the Na2CO3-regulated OECs change will facilitate the PSII assembly and oxygen evolution.Besides,photosystem II subunit L (PsbL) and TL29 also participate in the assembly of the PSII complex [54,55].The phosphorylation of PsbL was induced and the phosphorylation of TL29 was inhibited in response to Na2CO3(Figure S8E).Although the phosphorylation of PsbL was also reported in Arabidopsis [23],their regulatory mechanisms are not known.
Activities of PSI and ATP synthase regulated by reverse phosphorylation of Na2CO3-responsive proteins
In contrast to PSII,PSI drew little attention due to difficulties in accurately measuring its activity [56].The abundance changes of several PSI proteins (e.g.,Lhca1 and Lhca5) and Na2CO3-enhanced phosphorylation of Lhca2 and Lhca4 in alkaligrass allow the regulation of light absorption through the antenna modulation to prevent PSI damage (Figure S8C)[25].The STN7-regulated phosphorylation of Lhca4 was also induced in Arabidopsis when the plastoquinone overly reduced [57].Therefore,the enhancement of Lhca4 phosphorylation would be favorable for trapping and dissipation of excitations,working as a photoprotective mechanism of PSI [58].
Among the PSI core proteins,PsaA,PsaB,and PsaC [59]were decreased in alkaligrass (Figure S8C),barley (Hordeumvulgare) [60],and other halophytes [33,61].The phosphorylation of PsaC was enhanced in alkaligrass at 24 HAT (Figure S8C),which has been reported in Arabidopsis,green algae (Chlamydomonasreinhardtii),and spikemoss (Selaginellamoellendorffii) [23].The decreased abundances of PsaA,PsaB,and PsaC imply that saline-alkali stress inhibited the energy transfer of PSI.In addition,PsaD and PsaE provide docking sites for ferredoxin,and PsaF is important for interaction with the lumenal electron donor plastocyanin,while PsaG and PsaH participate in stabilizing PSI complex [62].In this study,these proteins were significantly accumulated,and the phosphorylation of PsaD,PsaE,and PsaH were also enhanced at 24 HAT of Na2CO3(Figure S8C),although onlyPsaHgene transcription was obviously induced at 24 HAT.Their phosphorylation events have been reported in Arabidopsis,green algae,andSynechocystis6803 [23],and the phosphorylation of PsaD was supposed to regulate the electron transfer from PSI to the electron acceptors in Arabidopsis chloroplast stroma [63].Therefore,we suggested that the salinity-/alkaliincreased abundances or phosphorylation of these proteins facilitate stabilization of the PSI complex (thus protecting it from photodamage),and activate the CET around PSI [62].
The gene expression and protein abundance of different subunits of ATP synthase were altered by Na2CO3stress in alkaligrass (Figure S7) and other plant species [4].The activity of ATP synthase is salinity-responsive,being regulated by the reversible protein phosphorylation [4,64].Several phosphorylation sites in ATP synthase subunits (α,β,δ,and ε) of spinach (Spinaciaoleracea) chloroplasts have been reported [64].In this study,Na2CO3inhibited the phosphorylation of α (Ser125),β (Ser497 and Thr52),and ε subunit (Thr110),but enhanced the phosphorylation of α (Ser9,Ser21,and Thr43) and β subunits (Thr53 and Ser445) in alkaligrass at 24 HAT (Figure S8C).This implies that the stability and rotation of F1 head of ATP synthase are modulated for the dynamic regulation of its activity to cope with alkali stress.
Different ROS homeostasis pathways employed in chloroplasts and other subcellular locations to cope with Na2CO3stress
Na2CO3treatment disrupted the electron transport in chloroplasts,as well as tricarboxylic acid cycle and respiration chain in mitochondria (Figures 2 and S8F),leading to the increases of H2O2and O2-in leaves [65].A previous proteomic study reported that many ROS-scavenging enzymes were altered in salinity-stressed leaves [4].Our results indicated that parts of AsA-GSH cycle (i.e.,APX and GPX pathways) were induced in leaves,but most ROS scavenging pathways in chloroplasts were inhibited,except for the APX pathway and SOD pathway (Figure 4).This implies that different pathways are employed in chloroplasts and other subcellular locations in leaves to cope with the short-term Na2CO3stress (12 h).While under longterm NaCl or Na2CO3stress (7 d),the pathways of SOD,POD,and CAT were all induced in leaves of alkaligrass [24,25].
Various non-enzymatic antioxidants are important for ROS scavenging [66].In this study,although the balance of AsA and DHA in chloroplasts was perturbed,the contents of AsA and DHA in leaves were stable.Additionally,the ratio of GSH/GSSG was stable in chloroplasts,and increased in leaves at 24 HAT (Figure 4).Furthermore,Na2CO3also increased glyoxalase I,but decreased chloroplast-localized cysteine synthase.Both enzymes are involved in GSH/GSSG balance (Figure S8F).All these imply that the AsAGSH cycle is inhibited in chloroplasts,but enhanced in other organelles and cytoplasm of leaf cells to cope with the alkali stress.In addition,a chloroplast-localized activator of bc1 complex kinase increased at 24 HAT,which would facilitate tocopherol cyclase phosphorylation to stabilize it at plastoglobules for vitamin E synthesis [67].The thylakoid membrane-localized vitamin E is a lipid antioxidant.This result is consistent with our previous finding of NaClincreased vitamin E content and tocopherol cyclase abundance in leaves of alkaligrass [25].
Additionally,the atlas of Na2CO3-responsive proteins indicated that modulation of Chl synthesis (Figure S8G),chloroplast movement and stability (Figure S8H) were critical for alkali adaptation,and ABA-dependent alkali-responsive pathways were employed to regulate both nuclear and chloroplastic gene expression and protein processes for osmoprotectant synthesis and signaling pathways in alkaligrass (Figure S8I and J).
Conclusion
Although NaCl-responsive mechanisms have been well-studied in various halophytes using proteomics approaches [4],the Na2CO3-responsive proteins and corresponding regulatory mechanisms in halophytes were rarely explored.This study is the first detailed investigation of Na2CO3-responsive proteins in chloroplasts using proteomics and phosphoproteomics approaches,which revealed several crucial Na2CO3-responsive pathways in halophyte chloroplasts (Figure S8).Our study showed that maintenance of energy balance between PSII and PSI,efficiency of PSII damage repair,cyclic electron transport,dynamic thylakoid membrane architecture,as well as osmotic and ROS homeostasis were essential for photosynthetic modulation in response to Na2CO3.Both the nuclearand chloroplast-encoded proteins were critical for the Na2CO3-responsive chloroplast function.More importantly,the newly-identified protein phosphorylation sites suggest that the reversible protein phosphorylation is important for regulating multiple signaling and metabolic pathways in chloroplasts to cope with the Na2CO3stress.Some of these Na2CO3-responsive proteins and phosphoproteins are potential saline-alkali stress biomarkers for further functional characterization and biotechnological application.
Materials and methods
Plant material treatment and biomass analysis
Seeds of alkaligrass [Puccinellia tenuiflora(Turcz.) scribn.et Merr.]were sowed on vermiculite and grown in Hoagland solution in pots under fluorescent light (220 μmol m-2s-1,12 h day and 12 h night) at 25 °C day and 20 °C night,and 75% humidity for 50 days.Seedlings were treated with 0 mM,150 mM,and 200 mM Na2CO3(pH 11) for 12 h and 24 h,respectively(Figure S9).After treatment,leaves were harvested,either used immediately or stored at-80°C for experiments.Shoot length and leaf fresh weight of seedlings were immediately measured.Dry weight,relative water content,as well as ion contents of K+,Na+,Ca2+,and Mg2+were determined according to the method of Zhao and the colleagues[26].
Membrane integrity,osmolytes,and ABA analysis
The malondialdehyde content and relative electrolyte leakage were determined using previous methods [68,69].Free proline and total soluble sugar contents were quantified with a spectrometer at 520 nm and 630 nm,respectively[26].The content of endogenous ABA was measured by an indirect ELISA method [70].
Photosynthesis and chloroplast ultrastructure analysis
Photosynthesis and Chl fluorescence parameters were measured using previous methods[25,69].Net photosynthetic rate,stomatal conductance,and transpiration rate were determined at 10:00 a.m.using a portable photosynthesis system LI-COR 6400 (LI-COR Inc.,Lincoln,Nebraska,USA).Chl fluorescence parameters were recorded using a pulse-amplitudemodulated (PAM) Chl fluorometer (Dua-PAM-100) (Heintz Walz,Effeltrich,Germany) and an emitter-detector-cuvette assembly with a unit 101ED (ED-101US).For the rapid fluorescence induction kinetics analysis,the OJIP were measured at room temperature (25 °C) with a portable fluorometer PAM-2500 (Walz,Effeltrich,Germany).The fluorescence measurement and calculation were performed according to the protocol of Strasser and the colleagues [71].Chl contents were determined according to the method of Wang and the colleagues [69].The ultrastructure of chloroplasts were observed under a JEOL-2000 transmission electron microscope (JEOL,Tokyo,Japan) according to Suo and the colleagues [72].
Measurements of ROS and antioxidant contents,and antioxidant enzyme activities
The O2-generation rate and H2O2content were measured according to Zhao and the colleagues[26].Reduced AsA,total AsA,GSSG,and total GSH contents were determined according to methods of Law and the colleagues[73].The activities of SOD,POD,CAT,APX,GR,and GST were measured as previously described [25,26].The activities of MDHAR,DHAR,and GPX were assayed according to Zhao and the colleagues[26].
Chloroplast isolation,protein extraction and purity assessment
Intact chloroplasts were isolated according to Ni and the colleagues[74],and the chloroplast protein for iTRAQ and phosphoproteomics analysis was extracted according to Wang and the colleagues [75].The purity of chloroplast protein was assessed by Western blot analysis with antibodies against marker proteins for different subcellular compartments according to Dai and the colleagues[76].Primary antibodies against marker proteins and protein loading amounts were listed in Table S14.
Proteomic analysis chloroplasts and leaves
The protein samples were extracted,fractioned,lyophilized,and resuspended for MS/MS analysis according to Zhao and the colleagues [26].The peptides of control (0 mM Na2CO3),12 HAT150,12 HAT200,24 HAT150,and 24 HAT200 were labeled with iTRAQ reagents 116,117,118,119,and 121(AB Sciex Inc.,Foster City,CA,USA),respectively.Three biological replicates were performed.The LC-MS/MS analysis was performed by Triple TOFTM5600 LC-MS/MS (AB Sciex Inc.,Concord,Canada) according to Zhao and the colleagues[26].The MS/MS data were submitted to database searching using the Paragon algorithm of ProteinPilot (version 4.0,AB Sciex Inc.) and the proteomics data was deposited to the Proteomics Identifications (PRIDE) database [77].The databases used were the UniProt Liliopsida database (320,685 entries) and the National Center for Biotechnology Information (NCBI) non-redundant database (7,262,093 sequences).
Total protein samples from leaves were prepared and analyzed using two-dimensional gel electrophoresis according to the method of Suo et al [72]and Dai and the colleagues [76].The MS and MS/MS spectra were acquired on a MALDITOF/TOF mass spectrometer (AB Sciex Inc.) [69].The MS/MS spectra were searched against the NCBI non-redundant green plant database (http://www.ncbi.nlm.nih.gov/)(3,082,218 sequences) using the search engine Mascot (version 2.3.0,Matrix Science,London,UK) (http://www.matrixscience.com) according to Meng and the colleagues [78].
Phosphoproteomic analysis of proteins from chloroplasts and leaves
After digestion,the chloroplast peptide samples of control,24 HAT150,and 24 HAT200 were labeled with stable isotope dimethyl labeling in light,intermediate,and heavy,respectively,according to the method of Boersema and the colleagues [79].In addition,the peptides from each leaf protein sample were labeled with iTRAQ reagents (113 and 116 for control,114 and 117 for 24 HAT150,as well as 115 and 119 for 24 HAT200) according to the manufacturer’s instructions,respectively (AB Sciex Inc.).The phosphopeptides were enriched using TiO2micro-column [80].LC-MS/MS analysis was performed using a nanoAcquity ultraperformance LC(Waters,Milford,MA,USA) coupled with an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific,Watham,MA,USA) [81].
For database searching,raw data files from chloroplast phosphoproteome were processed using Mascot server (version 2.3.0,Matrix Science) and searched against the NCBI green plant database (7,262,093 sequences) using an in-house Mascot Daemon (version 2.4,Matrix Science) [81].For phosphopeptide relative quantification,Mascot Distiller (version 2.5.1.0,Matrix Science) was used.Precursor ion protocol was used for peptide quantification and the ratios were calculated using the peak areas of extracted ion chromatograms based on the trapezium integration method [82].For leaf phosphoproteins,MS/MS data and peak lists were extracted using ProteinPilot (version 4.0,AB Sciex Inc.),searched against the NCBI green database (7,262,093 sequences) at a 95% confidence interval (unused ProtScore >1.3).After database searching,reliable quantification of individual phosphopeptide was achieved by the mean ± S.D.of triplicate experiments,the peptides with more than 1.5-fold changes in at least two replicates were considered to be changed at phosphorylation level.
Protein classification,hierarchical clustering,subcellular location prediction,and 3D structure analysis
Functional domains of proteins were analyzed using BLAST programs (http://www.ncbi.nlm.nih.gov/BLAST/),and then were clustered by cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm).The prediction of protein subcellular location was determined using five internet tools:YLoc (http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi),LocTree3 (https://rostlab.org/services/loctree3/),Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/),ngLOC (http://genome.unmc.edu/ngLOC/index.html),and ChloroP (http://www.cbs.dtu.dk/services/ChloroP/).The SWISS-MODEL comparative protein modeling server (http://swissmodel.expasy.org/) was employed to generate 3D structural models of phosphoproteins [83].
qRT-PCR analysis of homologous gene expression
Total RNA was isolated from leaves using the pBiozol plant total RNA extraction reagent (BioFlux,Hangzhou,China).A first-strand cDNA was obtained from 1 μg of total RNA using a PrimeScript?RT reagent kit (Takara Bio,Inc.,Otsu,Japan).The sequences of candidate genes were obtained from the local alkaligrass EST database using a BLASTn program.qRT-PCR amplification was performed using the specific primer pairs (Table S13) on a 7500 real time PCR system(Applied Biosystems Inc.,USA).The amplification process was performed according the method of Suo and the colleagues [72].
Western blot analysis
Western blotting was conducted according to Dai and the colleagues [76].The primary antibodies were raised in rabbits against the Arabidopsis PsbS,Cytf,D1,PsbO,PsaD,RBL,PGK,and SBPase.Signals were detected with ECL PlusTMreagent (GE Healthcare) according to the manufacturer’s instruction.Relative abundances were analyzed using the Image Master 2D Platinum Software (version 5.0,GE Healthcare).For the immunodetection of the FBA level in the WT andOE-PtFBA Synechocystis6803,antibodies against FBA and RBL was used,15 μg total protein aliquots of WT andOE-PtFBA(including the indicated serial dilutions) were loaded,and RBL was used as a loading control.
Overexpression of PtFBA in Synechocystis 6803
Full length cDNA ofPtFBAwas amplified by PCR using appropriate primers (Table S15).The PpsbAIIexpression vector was used to generateOE-PtFBAstrain.A fragment containing thePtFBAgene was amplified by PCR (Table S15)and then inserted intoNdeI sites of PpsbAIIto form the PpsbAII-PtFBAexpression vector construct,which was used to transform the WTSynechocystis6803 using a natural transfer method[84].The transformants were spread on BG-11 agar plates containing 10 μg ml-1of spectinomycin,then incubated in 2% (v/v) CO2in air and illumination at 40 μmol photons m-2s-1.TheOE-PtFBAcells in the transformants was segregated to homogeneity (by successive streak purification) as determined by PCR amplification(Table S15),reverse transcription (RT-PCR) analysis(Table S15),and immunoblotting [85].Cell growth and Chlacontent analysis were conducted according to Gao and the colleagues [86].
Statistical analysis
All the results are presented as means ± S.D.of at least three replicates.The physiological and proteomics data were analyzed by Student’sttest.P<0.05 was considered statistically significant.
Data availability
The chloroplast and leaf proteomics data were available in the Proteomics Identifications Database(PRIDE:PXD005491 for chloroplast and PXD005455 for leaf).The chloroplast and leaf phosphoproteomics data also have been deposited to the Proteomics Identifications Database (PRIDE:PXD005472 for chloroplast and PXD005471 for leaf).
CRediT author statement
Jinwei Suo:Methodology,Investigation,Writing -original draft.Heng Zhang:Methodology,Investigation,Formal analysis,Visualization.Qi Zhao:Visualization,Data curation.Nan Zhang:Investigation,Validation.YongxueZhang:Investigation,Formal analysis,Validation.Ying Li:Formal analysis,Data curation.Baohua Song:Methodology,Investigation.Juanjuan Yu:Data curation,Validation.Jianguo Cao:Investigation.Tai Wang:Resources,Writing -review &editing.Ji Luo:Software,Investigation.Lihai Guo:Methodology,Software.Jun Ma:Methodology,Software,Data curation.Xumin Zhang:Methodology,Resources,Data curation.Yimin She:Resources,Methodology.Lianwei Peng:Resources,Visualization,Investigation.Weimin Ma:Resources,Methodology,Visualization.SiyiGuo:Supervision,Writing -review &editing.Yuchen Miao:Resources,Writing -review &editing.Sixue Chen:Writing -review &editing.Zhi Qin:Conceptualization,Supervision,Resources,Project administration,Writing -review &editing.Shaojun Dai:Conceptualization,Methodology,Supervision,Funding,Project administration,Writing -review &editing.All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
The project was supported by The Foundation of Shanghai Science and Technology Committee (Grant No.17391900600),The Program for Professor of Special Appointment (Eastern Scholar) from The Shanghai Bureau of Higher Education (2011 and 2017),and The Natural and Science Foundation of Heilongjiang Provence (Grant No.ZD2019C003) to Shaojun Dai,as well as The Fund of Shanghai Engineering Research Center of Plant Germplasm Resources (Grant No.17DZ2252700).Peter Scott from University of Florida is acknowledged for critical reading and editing of the manuscript.
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2018.10.011.
ORCID
0000-0002-7375-2217 (Jinwei Suo)
0000-0002-7797-0430 (Heng Zhang)
0000-0003-0826-6068 (Qi Zhao)
0000-0002-3595-7553 (Nan Zhang)
0000-0003-3661-3063 (Yongxue Zhang)
0000-0001-8912-3100 (Ying Li)
0000-0003-2262-7229 (Baohua Song)
0000-0002-7209-0645 (Juanjuan Yu)
0000-0001-5732-1903 (Jianguo Cao)
0000-0003-2752-697X (Tai Wang)
0000-0002-5805-4787 (Ji Luo)
0000-0003-4093-189X (Lihai Guo)
0000-0003-2216-5714 (Jun Ma)
0000-0002-2810-6363 (Xumin Zhang)
0000-0002-0571-4630 (Yimin She)
0000-0002-2353-7500 (Lianwei Peng)
0000-0003-4964-415X (Weimin Ma)
0000-0003-2013-1772 (Siyi Guo)
0000-0002-4339-1238 (Yuchen Miao)
0000-0002-6690-7612 (Sixue Chen)
0000-0002-5611-3122 (Zhi Qin)
0000-0001-8063-6946 (Shaojun Dai)
 Genomics,Proteomics & Bioinformatics2020年3期
Genomics,Proteomics & Bioinformatics2020年3期
- Genomics,Proteomics & Bioinformatics的其它文章
- CircPlant:An Integrated Tool for circRNA Detection and Functional Prediction in Plants
- Genome Assembly and Pathway Analysis of Edible Mushroom Agrocybe cylindracea
- Paleo-polyploidization in Lycophytes
- Genome Size Evolution Mediated by Gypsy Retrotransposons in Brassicaceae
- Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles
- Characterization of Lysine Monomethylome and Methyltransferase in Model Cyanobacterium Synechocystis sp.PCC 6803
