Post-transcriptional mechanisms controlling neurogenesis and direct neuronal reprogramming
Elsa Papadimitriou, Dimitra Thomaidou
Abstract Neurogenesis is a tightly regulated process in time and space both in the developing embryo and in adult neurogenic niches.A drastic change in the transcriptome and proteome of radial glial cells or neural stem cells towards the neuronal state is achieved due to sophisticated mechanisms of epigenetic, transcriptional, and post-transcriptional regulation.Understanding these neurogenic mechanisms is of major importance, not only for shedding light on very complex and crucial developmental processes, but also for the identification of putative reprogramming factors, that harbor hierarchically central regulatory roles in the course of neurogenesis and bare thus the capacity to drive direct reprogramming towards the neuronal fate.The major transcriptional programs that orchestrate the neurogenic process have been the focus of research for many years and key neurogenic transcription factors, as well as repressor complexes, have been identified and employed in direct reprogramming protocols to convert non-neuronal cells, into functional neurons.The posttranscriptional regulation of gene expression during nervous system development has emerged as another important and intricate regulatory layer, strongly contributing to the complexity of the mechanisms controlling neurogenesis and neuronal function.In particular, recent advances are highlighting the importance of specific RNA binding proteins that control major steps of mRNA life cycle during neurogenesis, such as alternative splicing, polyadenylation, stability, and translation.Apart from the RNA binding proteins, microRNAs, a class of small non-coding RNAs that block the translation of their target mRNAs, have also been shown to play crucial roles in all the stages of the neurogenic process, from neural stem/progenitor cell proliferation, neuronal differentiation and migration, to functional maturation.Here, we provide an overview of the most prominent posttranscriptional mechanisms mediated by RNA binding proteins and microRNAs during the neurogenic process, giving particular emphasis on the interplay of specific RNA binding proteins with neurogenic microRNAs.Taking under consideration that the molecular mechanisms of neurogenesis exert high similarity to the ones driving direct neuronal reprogramming, we also discuss the current advances in in vitro and in vivo direct neuronal reprogramming approaches that have employed microRNAs or RNA binding proteins as reprogramming factors, highlighting the so far known mechanisms of their reprogramming action.
Key Words: direct neuronal reprogramming; in vivo glia-to-neuron conversion; microRNAs;neurogenesis; post-transcriptional regulation; RNA binding proteins
Introduction
Post-transcriptional regulation of gene expression plays a crucial role during neurogenesis and neuronal maturation and is considered to contribute majorly to the great proteome diversity and functional complexity of the various neuronal subtypes.RNA binding proteins (RBPs) and non-coding RNAs,such as microRNAs (miRNAs), participate in intricate post-transcriptional mechanisms controlling every step of RNA metabolism, such as alternative splicing (AS) (Makeyev et al., 2007; Licatalosi et al., 2012; Li et al., 2015b),polyadenylation (Zhu et al., 2006; Licatalosi et al., 2008; Sena et al., 2021),RNA editing (Merkurjev et al., 2018; Chen et al., 2019), stability (Lou et al.,2014; Dai et al., 2015; Alkallas et al., 2017; Lu et al., 2021), translation (Li et al., 2017a; Otsuka et al., 2019) and localization (Racca et al., 2010; Colak et al., 2013; Merkurjev et al., 2018).During neurogenesis, RBPs, and miRNAs recognize specific sequences mostly in intronic regions and in 3′ UTRs of premature and mature mRNAs, promoting the down-regulation of stemnessrelated mRNAs through mRNA decay and translation inhibition mechanisms(Visvanathan et al., 2007; Santos et al., 2016; Zhang et al., 2016; Velasco et al., 2019) and the parallel upregulation of neuronal-specific mRNAs related to neuronal differentiation, migration, axonal growth and synaptic transmission through AS, mRNA stabilization and promotion of translation mechanisms (Li et al., 2014; Franzoni et al., 2015; Lin et al., 2016; Saito et al., 2019; Dell’Orco et al., 2020).
Direct reprogramming of non-neuronal cells to neurons using neurogenic transcription factors (TFs; Heinrich et al., 2010, 2014; Vierbuchen et al.,2010; Aravantinou-Fatorou et al., 2015; Pataskar et al., 2016; Pereira et al.,2017; Rivetti Di Val Cervo et al., 2017; Karow et al., 2018), miRNAs (Yoo et al., 2011; Victor et al., 2014; Abernathy et al., 2017; Birtele et al., 2019;Papadimitriou et al., 2023), and chemical compounds (Li et al., 2015a; Gao et al., 2017) orchestrates the neuronal conversion through molecular cascades that resemble to a large extent the endogenous neurogenic mechanisms.Neurogenic TFs with pioneer factor activity have been shown to open neuronal gene loci in heterochromatin regions and, along with lineagespecific TFs instruct subtype-specific neuronal conversion of fibroblasts or glial cells (Wapinski et al., 2013; Pataskar et al., 2016; Matsuda et al., 2019;P?un et al., 2023).Alternatively, instead of interfering with the epigenetic and transcriptional machinery as the initiating step of reprogramming,strategies that employ neurogenic miRNAs (Yoo et al., 2011; Victor et al.,2014; Abernathy et al., 2017; Birtele et al., 2019; Papadimitriou et al.,2023) or downregulate non-neuronal RBPs (Xue et al., 2013, 2016a; Qian et al., 2020; Zhou et al., 2020), induce reprogramming by initially impacting post-transcriptional mechanisms, which alter at a secondary level the transcriptional and epigenetic status of cells undergoing reprogramming(Abernathy et al., 2017).
In this review, we provide an overview of the most prominent posttranscriptional mechanisms controlling neurogenesis and neuronal differentiation, focusing on the major RBPs and neurogenic miRNAs that instruct the establishment of the neuronal post-transcriptional program.We also present the so far identified cross-talk mechanisms between RBPs and miRNAs, which highlight the antagonistic or synergistic involvement of RBPs in miRNA function, as well as the targeting of RBPs by miRNAs during neurogenesis.Finally, we present paradigms of reprogramming strategies that convert non-neuronal cells to induced-neurons (iNs) by manipulating miRNA- or RBP-controlled post-transcriptional mechanisms and discuss the current evidence regarding theirin vivoglia-to-neuron conversion capacity as a strategy for neuronal replacement in animal models of neurodegenerative diseases and CNS trauma.
Search Strategy
This review analyzes studies found on the PubMed database using the following keywords: RBPs/miRNAs and neurogenesis, direct neuronal reprogramming and RBPs/miRNAs,in vivodirect reprogramming and RBPs,in vivoglia to neuron conversion and RBPs/miRNAs, interplay of RBPs and miRNAs in neurogenesis, alternative splicing in neurogenesis.All studies were published between 1996 and 2023 and represent the most relevant advances in the field.
Regulation of the Neurogenic Program by RNA Binding Proteins
Control of neurogenic splicing programs by PTBP1 and PTBP2 during early neurogenesis
Neural stem/progenitor cells (NSPCs) express high levels of the global regulator of alternative pre-mRNA splicing, polypyrimidine tract binding protein 1 (PTBP1, also known as PTB and hnRNP I).Genome-wide mapping studies have shown that PTBP1 preferentially binds to pyrimidine-rich sequences in pre-mRNAs, and regulates alternative exon inclusion or skipping in a location-dependent manner.More specifically, when PTBP1 binds upstream or within an exon, it acts as a repressor, but when it binds only on the downstream side, it activates inclusion.PTBP1-repressed exons have longer and stronger polypyrimidine tracts relative to PTBP1-activated exons that tend to have weaker 5′ splice sites (Llorian et al., 2010).PTBP1 acts majorly by inhibiting neuronal specific isoforms in non-neuronal cells or NSPCs (Xue et al., 2013, 2016; Linares et al., 2015).
As NSPCs exit the cell cycle, PTBP1 expression is gradually reduced, allowing for the upregulation of its neurogenic paralog, polypyrimidine tract binding protein 2 (PTBP2, also known as nPTB), which is critical for the progression of neurogenesis, as it initiates a switch in the repertoire of splicing events favoring neuronal differentiation of the neuronal committed progenitors(Boutz et al., 2007).PTBP2 is essential for the proper splicing of various transcripts involved in neuronal function, such as the postsynaptic density protein 95 (Psd-95), gephyrin (Gphn), gamma-aminobutyric acid type A receptor subunit gamma2 (Gabrg2), and calcium/calmodulin-dependent protein kinase II alpha/beta (Camk2a/b) (Figure 1; Boutz et al., 2007;Licatalosi et al., 2012; Zheng et al., 2012; Li et al., 2014).Interestingly,Ptbp2mRNA itself is a major target for destabilization by PTBP1 (Boutz et al.,2007; Spellman et al., 2007; Zheng et al., 2012; Figure 1).More specifically,PTBP1 represses the inclusion of the alternative exon 10 inPtbp2pre-mRNA,creating a premature termination codon, which triggers degradation ofPtbp2transcript due to non-sense mediated decay (NMD; Boutz et al., 2007;Spellman et al., 2007; Zheng et al., 2012).
In accordance with their unique spatiotemporal expression during neurogenesis, PTBP1 and PTBP2 regulate overlapping, but also distinct repertoires of splicing events (Licatalosi et al., 2012; Zheng et al., 2012;Vuong et al., 2016).HITS-CLIP experiments enabling the identification of direct PTBP2-RNAin vivointeractions revealed that the major action of PTBP2 in the developing mouse is to inhibit splicing of alternative adult exons present in pre-mRNAs related to synaptogenesis and neuronal plasticity,tightly controlling the correct timing of neuronal maturation of differentiating progenitors (Licatalosi et al., 2012; Zheng et al., 2012).However, in later stages of neurogenesis, as newborn neurons mature, PTBP2 is again downregulated to allow a second transition in the neurogenic splicing program towards the expression of neuronal maturation transcripts, such asPsd-95present in excitatory synapses (Zheng et al., 2012).At earlier stages of neurogenesis,both PTBP1 and PTBP2 repress splicing ofPsd-95pre-mRNA alternative exon 18, leading to premature translation termination and degradation ofPsd-95transcript by NMD.The sequential loss first of PTBP1 and then of PTBP2, as neurogenesis progresses, allows the skipping of exon 18 and expression ofPsd-95during synaptogenesis (Zheng et al., 2012).These sequential changes in PTBP1 and PTBP2 expression define three phases of splicing regulation during neurogenesis, allowing NSPCs, differentiating neuronal progenitors,and newborn neurons undergoing synaptic maturation to be defined by different, tightly regulated splicing programs (Zheng et al., 2012).
Other well-studied targets of PTBP1 include the TF pre B cell leukemia homeobox 1 (Pbx1) related to motor neuron development (Linares et al.,2015) and the pro-apoptotic protein BCL2 antagonist/killer 1 (Bak1) (Lin et al., 2020; Figure 1).During ESC differentiation into NSPCs, PTBP1 was shown to affect the splicing of the TFs transcription factor 20 (Tcf20), mediator complex subunit 23 (Med23), GATA binding protein 2a (Gata2a) andPbx1(Linares et al., 2015).Further study of PBX1, revealed that PTBP1 controls an isoform switch fromPbx1bto its longer neuronal counterpartPbx1a.PBX1A is important for motor neuron development, possibly by increasing the expression ofHoxc5through the enhancement of MEIS1 recruitment (Linares et al., 2015).In a recent study, PTBP1 was also linked to the regulation of the apoptotic potency of NSPCs during cortical development by controlling the alternative splicing ofBak1pre-mRNA.Later in neurogenesis, downregulation of PTBP1 leads to degradation ofBak1via NMD, attenuating apoptosis competence of differentiating neurons, which is critical for neuronal survival(Lin et al., 2020).
Control of neurogenic post-transcriptional programs by NOVA, RBFOX and ELAVL proteins
Apart from the switch in the expression of PTBP1 towards the upregulation of PTBP2 along the progress of neurogenesis, other neuronal specific RBPs are also being gradually upregulated to foster the activity of the neuronal posttranscriptional program, such as the RNA binding fox-1 homolog (RBFOX)family RBFOX1/2/3, the neuro-oncological ventral antigen (NOVA) family NOVA1/2 and the embryonic-lethal abnormal vision-like (ELAVL) family(ELAVL1/2/3/4, also known as HuR/B/C/D; Figure 1).For detailed reviews on the role of RBPs in neurogenesis, we refer the reader to previous studies (Pilaz and Silver, 2015; Bhat et al., 2022).
Would the Lord banish7 him forever from the heaven of warmth and light? The kindest eyes that Tibley had ever seen smiled at him and gently a voice whispered
RBFOX family
It has been reported that ARE containing mRNAs tend to accumulate in the neural lineage and might undergo coordinated upregulation during neural differentiation (Dai et al., 2015).Indeed, several genes that are upregulated during neurogenesis contain AREs in their 3′UTRs, such asGap43(Kohn et al.,1996; Tsai et al., 1997), acetylcholinesterase (Ache) (Deschênes-Furry et al.,2003), the neuronal cytoskeleton markers, microtubule-associated protein tau (Mapt) (Aranda-Abreu et al., 1999), and tubulin beta 3 class III (Tubb3),the brain-enriched RBPs,Elavl2/3/4andNova1, and the components of neurotrophic pathways, brain-derived neurotrophic factor (Bdnf) andNtrk2(Dai et al., 2015; Figure 1).
Interestingly, the 3′UTR ofElavl4has been shown to be additionally targeted by miR-375, a miRNA that is downregulated during the late stages of cortical development.The decrease of miR-375 allows the de-repression of ELAVL4,leading to the subsequent enhancement of neurite outgrowth in developing neurons (Abdelmohsen et al., 2010).
For two weeks, Mark, military uniform and all, accompanied Susan to and from work each day. He taught her how to rely on her other senses, specifically her hearing, to determine where she was and how to adapt to her new environment. He helped her befriend the bus drivers who could watch out for her, and save her a seat.
The jackal felt it was quite hopeless to get what he wanted, and asked, Tell me, mother dove, how have you suddenly become so wise ? It was the heron who told me, replied she
Rbfox3knockdown (KD) in the developing chicken spinal cord was shown to inhibit the late neuronal differentiation of developing neurons irrespective of their subtype specificity, indicating that RBFOX3 affects the general differentiation program in neurons.Alternative splicing of NUMB endocytic adaptor protein (Numb) pre-mRNA by RBFOX3 appeared crucial for proper neuronal differentiation (Kim et al., 2013).Although NUMB has been related to the regulation of Notch signaling and asymmetric cell divisions (Petersen et al., 2006; Luo et al., 2020), the alternative exon 12 exclusion inNumbpre-mRNA by RBFOX3 probably has an additional role in the regulation of migration of developing neurons (Kim et al., 2013).Studies inRbfox3KO mice highlighted its role in hippocampal circuit balance and function, by regulating synaptic transmission, dendritic complexity, spine density, and plasticity(Wang et al., 2015b).Additionally, RBFOX3 was further shown to affect adult hippocampal neurogenesis and hippocampal synaptic transmission, by regulating exon usage of many identified synaptic genes (Lin et al., 2016).
The increased expression of RBFOX1 and RBFOX3 in the adult mouse cortex in comparison to the embryonic cortex ensures their predominance over the developmentally controlled splicing of their targets, thus promoting the switch to the adult splicing program (Weyn-Vanhentenryck et al., 2014).Along this,recent studies have identified neuronal maturation mRNAs that are alternative spliced by RBFOX1, such as the vesicle-associated membrane protein (Vamp),shown to play central role in excitatory/inhibitory synaptic balance (Vuong et al., 2018) and the neurotrophic receptor tyrosine kinase 2 (Ntrk2, also known asTrkB) isoformTrkB.T1with prominent role in hippocampal brainderived neurotrophic factor-dependent long term potentiation (Tomassoni-Ardori et al., 2019).Additionally, the study of a tripleRbfoxKO mouse model highlighted the cytoskeletal protein ankyrin G (ANKG) as an “interaction hub”for the assembly of the axon initial segment during axonal growth (Jacko et al., 2018; Figure 1).
Nonsense mediated decay (NMD) is a conserved RNA degradation mechanism originally identified as a quality control pathway.This pathway rapidly degrades mRNAs harboring premature termination codons and is mediated by UPF1 RNA helicase and ATPase (UPF1), which is essential for NMD, along with the adapter proteins, UPF2 regulator of nonsense mediated mRNA decay (UPF2) and UPF3B regulator of nonsense mediated mRNA decay (UPF3B; reviewed in Wei-Lin Popp and Maquat, 2013; Lykke-Andersen and Jensen, 2015).More recent studies have shown that NMD is also a regulatory pathway necessary for developmental processes, such as brain development (for a recent review see (Jaffrey and Wilkinson, 2018).NMD has been proven to be an additional post-transcriptional mechanism controlling the transition from pluripotent to differentiated cell state.Indeed, UPF1 was shown to promote NSPCs proliferation by selectively targeting mRNAs encoding G1/S inhibitor proteins, such asCdkn1Aand the cyclin-dependent kinase inhibitor 1B (Cdkn1B, also known asp27).Further on, UPF1 was found to support TGF-β signaling activation by repressing the TGF-β inhibitor SMAD family member 7 (SMAD7), thereby preventing neuronal differentiation (Lou et al., 2014; Figure 1).The NMD pathway components UPF3B and UPF1 become progressively decreased as neurogenesis proceeds, resulting in the upregulation of transcripts important for neural development and function(Bruno et al., 2011; Jolly et al., 2013; Lou et al., 2014; Alrahbeni et al., 2015).Interestingly, the role of UPF3B downregulation has been proposed to be critical for proper neurogenesis and neuronal maturation, corroborating the relation ofUpf3Bmutations with neurodevelopmental disorders, such as intellectual disability and autism (Laumonnier et al., 2010; Jolly et al., 2013;Nguyen et al., 2013; Alrahbeni et al., 2015).Although functioning at lower levels compared to NSPCs, the NMD machinery is still necessary for proper neuronal function and has been reported to play a role in axon guidance and synaptic plasticity (Giorgi et al., 2007; Colak et al., 2013).
NOVA family
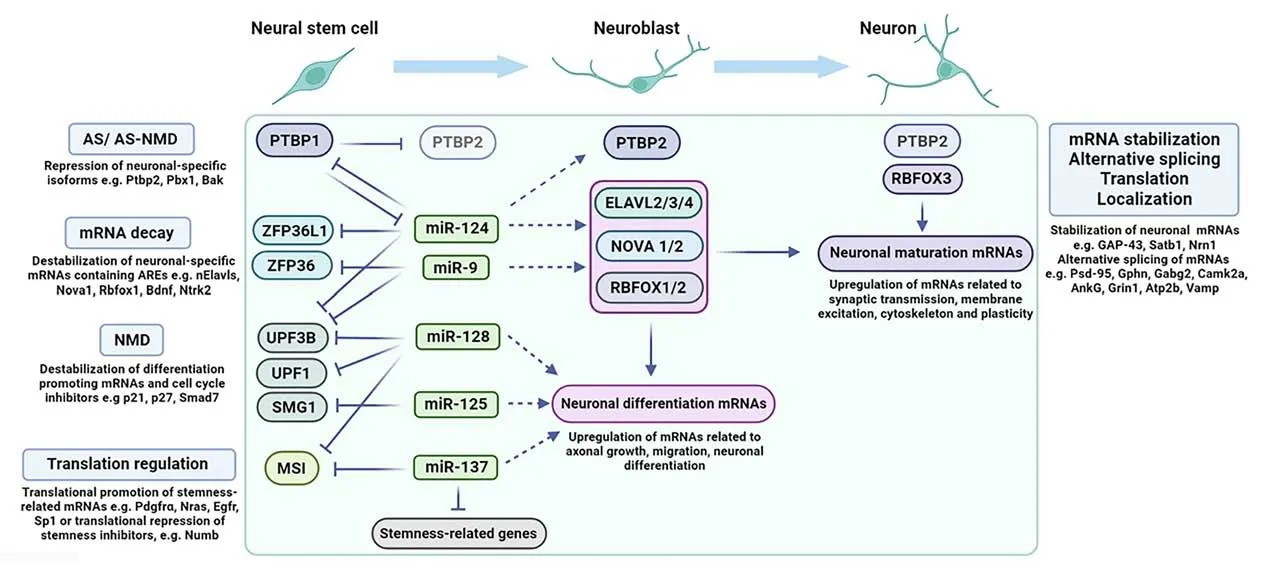
Figure 1 |Post-transcriptional mechanisms mediated by RBPs and miRNAs orchestrating neurogenesis.Post-transcriptional mechanisms controlling neurogenesis that are discussed in the text, giving particular emphasis on the role of neurogenic miRNAs in regulating RBPs expressed in neural stem/progenitor cells.Early in the course of neurogenesis the neurogenic miRNAs that are gradually upregulated, miR-124, miR-9, miR-128, miR-125, and miR-137 target for downregulation RBPs that inhibit the expression of neuronal specific mRNAs by (a) alternative splicing (AS) or alternative splicing coupled with nonsense mediated decay (AS-NMD),such as Ptbp1, (b) mRNA decay mechanisms, such as Zfp36l1 and Zfp36, (c) nonsense mediated decay (NMD), such as Upf1 and Upf3b and the kinase Smg1 that phosphorylates UPF1 and (d) translation inhibition, such as Msi.Through these actions, these neurogenic miRNAs indirectly de-repress numerous neurogenic mRNAs, such as Ptbp2, the neuronal RBPs Elavl2/3/4, Nova1/2, Rbfox1/2 and other neuronal-specific mRNAs related to axonal growth, migration, and differentiation.As discussed in the text, miR-137, apart from directly targeting Msi, also competes with MSI in directly targeting stemness-related mRNAs.Later in neurogenesis, PTBP2 levels decline (shown in lighter blue color) allowing for adultspecific alternative splicing to occur.In parallel, the increase of RBFOX3 in synergy with other neuronal specific RBPs promotes the upregulation of neuronal maturation mRNAs related to synaptic transmission, membrane excitation, cytoskeleton, and plasticity by post-transcriptional mechanisms, including (a) stabilization, (b) alternative splicing, (c) promotion of translation and (d) transport and control of local translation.Created with BioRender.com.ANKG: Ankyrin G; AREs: AU-rich elements; AS: alternative splicing; AS-NMD: alternative splicing coupled with nonsense mediated decay; ATP2B: ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2; BAK: BCL2 antagonist/killer 1; BDNF: brain-derived neurotrophic factor; CAMK2A: calcium/calmodulin-dependent protein kinase II alpha; EGFR: epidermal growth factor receptor; ELAVL2/3/4: embryonic lethal abnormal vision-like 2/3/4; GABRG2: gamma-aminobutyric acid type A receptor subunit gamma2; GAP-43: growth associated protein 43; GPHN: gephyrin; GRIN1: glutamate ionotropic receptor NMDA type subunit 1; MSI: musashi; nELAVLs: neuronal ELAVLs; NOVA1/2: neuro-oncological ventral antigen 1/2; NRAS: Neuroblastoma Ras proto-oncogene; NRN1: neuritin 1; NTRK2:neurotrophic receptor tyrosine kinase 2; NUMB: NUMB endocytic adaptor protein; PBX1: pre B cell leukemia homeobox 1; PDGFRA: platelet-derived growth factor receptor alpha;PSD-95: postsynaptic density protein 95; PTBP1/2: polypyrimidine tract binding protein 1/2; RBFOX1/2/3: RNA binding fox-1 homolog 1/2/3; SATB1: Special AT-Rich Sequence Binding Protein 1; SMAD7: SMAD family member 7; SMG1: SMG1 nonsense mediated mRNA decay associated PI3K related kinase; SP1: SP1 transcription factor; UPF1: UPF1 RNA helicase and ATPase; UPF3B: UPF3B regulator of nonsense mediated mRNA decay; VAMP: vesicle-associated membrane protein; ZFP36: ZFP36 ring finger protein; ZFP36L1: ZFP36 ring finger protein like 1.
NOVA1 and NOVA2 RBPs bind directly to RNA sequences harboring YCAY motifs and both regulate alternative splicing depending on the position of their binding to pre-RNAs, similar to the mode of action of RBFOX proteins.Despite their high homology and the identification of ~700 NOVA1/2 targeted alternative exons, these two RBPs have distinct and even reciprocal expression patterns in the mouse brain (Zhang et al., 2010).NOVA2 is expressed at high levels in the cortex and hippocampus, and lower levels in the midbrain and spinal cord, whereas NOVA1 is expressed, oppositely,at high levels in the midbrain and spinal cord and lower levels in the cortex(Saito et al., 2016).NOVA1- and NOVA2-specific HITS-CLIP experiments combined with transcriptomic data fromNova1KO orNova2KO mice have allowed the identification of common as well as unique roles for these two RBPs during brain development (Ule et al., 2005; Zhang et al., 2010; Saito et al., 2016, 2019).NOVA1/2 possesses a central role in the orchestration of neuronal-specific alternative splicing during neuronal differentiation, majorly regulating the alternative splicing of synaptic genes (Ule et al., 2005; Zhang et al., 2010).Interestingly, in the mouse brain about half of NOVA1/2 target transcripts encode phosphoproteins, and many microexons regulated by NOVA1/2 introduce phosphorylation sites, suggesting that NOVA1/2 directly affect the phosphorylation patterns of brain proteins modulating in this way downstream protein-protein interactions and physiological functions(Zhang et al., 2010).NOVA2 paralog, specifically, is indispensable for proper cortical lamination, controlling the alternative splicing of the Reelin pathway component, DAB adaptor protein 1 (Dab1), and plays a crucial role in the migration of late-born neurons (Yano et al., 2010).Additionally, NOVA2 has been identified to uniquely regulate alternative splicing of a set of transcripts encoding key components in cortical, brainstem, and spinal axon guidance/outgrowth pathways during neuronal differentiation (Saito et al., 2016).Recently, NOVA2 was found to mediate alternative splicing diversity across different brain regions, as well as different neuronal subtypes within the same region.For example, differential NOVA2-dependent alternative splicing of inositol 1,4,5-trisphosphate receptor type 1 (Itrp1) andDab1transcripts has been observed in excitatory versus inhibitory cortical neurons, possibly due to the difference in the RBPs’ expression ratio in each subtype (Saito et al., 2019).Inference of NOVA splicing-regulatory network after extensive bioinformatic data integration from HITS-CLIP and microarray experiments performed inNovaKO brains, revealed that 15% of identified NOVA splicing targets were under NOVA and RBFOX combinatorial control (Zhang et al.,2010).Mouse brain studies have indicated that NOVA1/2 proteins might also have regulatory functions in the cytoplasm.CLIP experiments in the mouse brain indicated that NOVA can bind to the 3′UTR of target mRNAs and regulate alternative polyadenylation (Licatalosi et al., 2008).Additionally,NOVA was found to colocalize with target mRNAs, including glycine receptor alpha 2 (Glra2) and G-protein-activated inward rectifying potassium type 2(Girk2), in dendrites of spinal cord motor neurons (Racca et al., 2010).
ELAVL family
The ELAVL family consists of four highly homologous proteins that bind to U- and AU-rich elements (AREs).ELAVL1 (also known as HuR) is expressed in a wide range of non-neuronal cells, while ELAVL2/3/4 (also known as HuB/C/D) exhibit neuronal specific expression and are therefore referred to neuronal ELAVLs (nELAVLs) (Fan and Steitz, 1998; Hinman and Lou, 2008).Although ELAVL1 has been considered a ubiquitous cytoplasmic RBP with a largely undefined role in neurogenesis, it has been shown to stabilize deltalike canonical Notch ligand 1 (Dll1) mRNA in neuroepithelial cells undergoing mitosis, supporting its role in the proliferation of neural progenitors (García-Domínguez et al., 2011).Additionally, ELAVL1 was found to regulate the association of functionally related mRNAs and proteins in actively translating polysomes during neocortical development in a stage-specific manner, and its absence disrupted correct postnatal neocortical lamination (Kraushar et al., 2014).A recent study demonstrated that developmental cytoplasmicto-nuclear translocation of ELAVL1 is essential for adult neurogenesis in the hippocampus, where it controls the alternative splicing of a plethora of transcripts.Among these transcripts, alternative splicing of focal adhesion kinase (Fak) pre-mRNA by ELAVL1 was found to tightly control its levels and activity, which is crucial for proper neuronal migration and development (Wang et al., 2019).
The nELAVLs have been primarily related to mRNA stabilization through their binding to AREs in the 3′UTRs of their target mRNAs in the cytoplasm,however today it is well-established that the nELAVLs are pleiotropic RBPs implicated also in pre-mRNA splicing (Ince-Dunn et al., 2012) and alternative polyadenylation in the nucleus (Sena et al., 2021).Indeed, nELAVL-HITS-CLIP experiments performed in mouse brains combined with microarray data fromElavl3/4double KO mice, demonstrated that the nELAVLs bind majorly on the 3′UTRs and secondarily on intronic regions of target pre-mRNAs in the brain, indicating two apparently independent functions of nELAVL-RNA interactions, which however intersect in the control of the synthesis of the major excitatory neurotransmitter glutamate (Ince-Dunn et al., 2012).Further nELAVL-CLIP experiments combined with RNA-seq data from the human brain identified numerous novel nELAVL targets with important roles in neuronal function, shown to be regulated either by alternative splicing or stabilization in the cytoplasm.Importantly, some of these targets were linked to neurodegenerative diseases such as AD (Scheckel et al., 2016).Interestingly,human nELAVLs were for the first time shown to bind to a class of non-coding RNAs, termed Y RNAs, which sequester nELAVLs from their mRNA targets during stress conditions, with possible implication in the pathogenesis of AD(Scheckel et al., 2016).
ELAVL4 is the most well-studied member of the nELAVL family and has been shown to be upregulated early in the course neurogenesis (Akamatsu et al.,2005; DeBoer et al., 2014; Popovitchenko et al., 2020), while its expression decreases, but persists in the glutamatergic neurons of the adult deep neocortical layers and the hippocampus (DeBoer et al., 2014).ELAVL4 resides mostly in the cytoplasm, often in the dendrites of developing neurons, where it stabilizes the levels and enhances the translation of its target mRNAs, many of which are implicated in neuronal differentiation, neurite outgrowth, axonal elongation, cytoskeleton organization, and neuronal plasticity (Bolognani et al., 2009).Prominent targets of ELAVL4 are the growth-associated protein 43 (Gap43) which plays a crucial role in neurite outgrowth during neuronal development and plasticity (Anderson et al., 2001; Pascale et al., 2004),neuritin 1 (Nrn1, also known asCpg15), involved in axonal elongation during neuronal differentiation and axonal injury (Akten et al., 2011; Gomes et al.,2017) and the cortical development TF special AT-rich sequence binding protein 1 (Satb1; Wang et al., 2015a; Figure 1).More recent studies unveiled a more versatile role for ELAVL4, not only in the control of alternative splicing and alternative polyadenylation of neuronal transcripts (Zhu et al., 2006;Bellavia et al., 2007), but also in the regulation of non-coding RNAs such as circular RNAs and miRNAs (Dell’Orco et al., 2020, 2021; Zimmerman et al.,2020).Interestingly, ELAVL4 was found to regulate the synaptic expression and transport of circular RNAs related to synaptic plasticity, such as the circHomer1a (Zimmerman et al., 2020), circCreb1 and circUpf2 (Dell’Orco et al., 2020), participating in competing endogenous RNA networks that regulate the expression of genes associated with brain development and remodeling of neuronal connections (Dell’Orco et al., 2021).
In a recent study, it was shown that ELAVL4 appears in distinct mRNA isoforms that differ mainly in their 5′UTR.Interestingly, the translation of these isoforms in radial glia progenitors and early neurons is controlled by the RBP,CUGBP Elav-like family member 1 (CELF1), which binds selectively to the 5’UTR of specificElavl4isoforms, inhibiting their translation.Importantly, CELF1 regulation ofElavl4translation dictates the development of glutamatergic neurons (Popovitchenko et al., 2020).
Ashliman continues, It is easy to see how a child, abused by the principal authority in her household-the individual who should be her most powerful protector-could see herself as being without hand, the human extension that must directly allow use to manipulate and control the world outside ourselves . In addition, mutilation to save one s virginity or to keep a vow62 has been a common theme in both the East and the West since ancient times (Zipes, Fairy Tale Tradition 506). Maria Warner points out, Only horribly disfigured in this way can she become inviolable and so resist (348).
Little is known about the specific functions of the other nELAVL members,ELAVL2 and ELAVL3.Recently,Elavl2KD followed by RNA-seq in human primary neurons revealed a role of ELAVL2 in affecting the circuitry of synaptic genes, genes associated with autism spectrum disorder, and alternatively spliced genes, while, interestingly, identified co-expression modules containing genes with conserved regulation downstream of both ELAVL2 and RBFOX1 (Berto et al., 2016).A role for ELAVL3 has been reported in the cerebellum, as Purkinje cells ofElavl3KO mice exhibited loss of neuronal polarity characterized by the leakage of somato-dendritic organelles into the axons.ELAVL3 was shown to control the alternative splicing ofAnkG,which is required for the generation of axon potentials in mature neurons, by excluding the embryonically expressed alternative exon 34 as neurons mature(Ogawa et al., 2018).
The Fairy finally made Narcissus promise that he would remain invisible when he was with the Princess, since she felt sure that this would make things easier for all of them
The role of RNA modifications in neurogenesis
RNA modifications, such as N6-methyladenosine (m6A), N1-methyladenosine,5-methylcytosine, 5-hydroxymethylcytosine, pseudouridine, inosine, uridine,and ribose-methylation, collectively referred to as the epitranscriptome, play an important role in gene expression regulation by affecting RNA metabolism(for a recent review see Zhou et al., 2020).m6A is the most abundant and well-studied RNA modification, implicated in pre-mRNA splicing (Xiao et al.,2016) and pri-miRNA processing (Alarcón et al., 2015a, b) in the nucleus,as well as in translational control (Li et al., 2017a; Slobodin et al., 2017) and mRNA decay (Du et al., 2016) in the cytoplasm.
Importantly, the developing and adult brain transcriptome is highly enriched in m6A modifications (recently reviewed in (Widagdo and Anggono, 2018;Livneh et al., 2019; Sokpor et al., 2021).A growing body of evidence indicates the importance of m6A modification in orchestrating neural development and function, including NSPC proliferation and differentiation (Li et al.,2018), synaptic transmission (Merkurjev et al., 2018), and neuronal plasticity(Widagdo et al., 2016).
The m6A machinery consists of three functional components namely, the m6A methyltransferases (“writers”), such as the methyltransferase-like protein 3 (METTL3) and METTL14, the m6A demethylases (“erasers”), such as fat mass and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase AlkB homolog 5 (ALKBH5), and m6A binding or interacting proteins (“readers”), such as YTH N6-methyladenosine RNA binding proteins,including YTHDC1–2, and YTHDF1–3, as well as eukaryotic translation initiation factor 3 (eIF3), insulin like growth factor 2 mRNA binding proteins 1–3 (IGF2BP1–3), ELAVL1 and fragile X messenger ribonucleoprotein 1 (FMR1,also known as FMRP) (for a recent review see Yang et al., 2018).
All this time Prince Narcissus, gloomy and despairing, was kept a prisoner by Melinette in her castle in the air, and in spite of all the splendour by which he was surrounded, and all the pleasures which he might have enjoyed, his one thought was to get back to Potentilla
In other reprogramming approaches that employed miRNAs, overexpression of miR-124 and miR-25, along with repression of let-7 in MG was shown to induceAscl1and efficiently convert mature MG into a neuronal/retinal progenitor cell phenotype.ScRNA-seq of reprogrammed MG confirmed the acquisition of a gene expression profile similar to retinal progenitor cells and retinal neurons and also revealed synergistic mechanisms between these miRNAs in targeting shared mRNAs, such as Krupper-like transcription factor (Klf4) and the dickkopf Wnt signaling pathway inhibitor 3 (Dkk3) or in cooperatively targeting different components of the REST complex, such asRest,Scp1andRcor1(Wohl et al., 2019).In another approach, miR-34b/c,identified in a microRNA-mRNA paired microarray screening to be among the most upregulated microRNAs during dopaminergic differentiation, was used along with ASCL1 and nuclear receptor subfamily 4, group A, member 2(NR4A2, also known as NURR1) and was shown to enhance their effectiveness to transdifferentiate fibroblasts into dopaminergic neurons (De Gregorio et al., 2018).
Decreasing m6A levels by eitherMettl14cKO orMettl3KD has been reported to prolong cell cycle progression in NSPCs and to delay the generation of upper-layer neurons in the mouse cortex (Yoon et al., 2017).Further comparison of m6A-seq data from human forebrain organoids and E13.5 mouse forebrains revealed many m6A-tagged transcripts that are conserved and related to neurogenesis and neuronal differentiation.Interestingly, this analysis further revealed that m6A mRNA methylation is more prevalent in human, while the human specific m6A-tagged transcripts were enriched for genes linked to mental disorders (Yoon et al., 2017).In another study,Mettl3deletion inhibited adult NSC proliferation, promoted adult NSCs’differentiation toward the glial lineage, and inhibited morphological maturation of newborn neurons bothin vitroandin vivo.Interestingly,Mettl3depletion reduced the levels of histone methyltransferase, enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and H3K27me3,uncovering a crosstalk between RNA methylation and histone modifications during neurogenesis (Chen et al., 2019).Additionally, the KO of the m6A“reader”Ythdf2caused a dramatic reduction in the overall cortical thickness ofYthdf2-deficient embryonic forebrains (Li et al., 2018).YTHDF2 is known to bind m6A-methylated mRNAs and promote mRNA decay (Du et al.,2016), thus upon loss ofYthdf2-mediated RNA degradation, an increase was observed in the expression of m6A-tagged transcripts associated with cortical neural development (Li et al., 2018).The m6A “eraser” FTO is expressed most prominently at later stages of neurogenesis and plays an important role in adult neurogenesis.Loss ofFtowas shown not only to reduce the proliferation of adult NSCs, but also to inhibit neuronal differentiation in both neurogenic regions of adult mice, leading to impaired learning and memory.Interestingly,FtoKO was found to dysregulate many genes of the brainderived neurotrophic factor pathway (Li et al., 2017b).Additionally, m6Aseq of adult mouse forebrain synaptosomes revealed that m6A is enriched in synaptic transcripts.Further on, KD of m6A readersYthdf1orYthdf13in hippocampal neurons caused aberrant spine morphology and synaptic dysfunction (Merkurjev et al., 2018), suggesting an important role for m6A readers at the synapses.
Regulation of mRNA Degradation Pathways in Neurogenesis
Competition of RBPs for the regulation of ARE containing mRNAs
ARE, which is most often located in the 3′ UTR of mRNAs, is the typical sequence responsible for controlling mRNA stability through its interaction with RBPs in the cytoplasm.AREs are contained in 5–8% of human mRNAs coding for proteins involved in various biological functions, such as cell communication, signal transduction, transcription, immune response,proliferation, and developmental processes, including neurogenesis (Bakheet et al., 2006).The RBPs that bind to AREs and destabilize their target mRNAs include, AU-rich element RNA binding protein 1 (AUF1), KH-type splicing regulatory protein (KHSRP, also known as KSRP), as well as the ZFP36 ring finger protein (ZFP36) family members, ZFP36, ZFP36L1, and ZFP36L2.In parallel, members of the ELAVL family, such as ELAVL1, act by stabilizing ARE containing mRNAs (for a recent review see Otsuka et al., 2019), often by antagonizing the action of destabilizing proteins (Chen et al., 2002;Hambardzumyan et al., 2009).Recently, it was reported that AREs are also present in introns being more abundant than previously thought, occurring in > 50% of human genes.The behavior of intronic AREs was quite recently proposed to be consistent with the preferential nuclear localization and translational enhancing ability of ELAVL1 (Bakheet et al., 2018).
The RBFOX family members RBFOX1/2/3 have been identified as global regulators of alternative splicing during neurogenesis, binding to the (U)GCAUG sequence in introns flanking alternative exons and enhancing or inhibiting alternative exon inclusion depending on the position of their binding to neurogenic pre-mRNAs (Weyn-Vanhentenryck et al., 2014).Expression of RBFOX1 begins early in cortical neurogenesis and persists in mature neurons of the adult cortex; RBFOX2 has a more prominent role in the development of the cerebellum, while RBFOX3 is expressed later as newborn neurons mature and prevails in the adult cortex (Gehman et al., 2011; Weyn-Vanhentenryck et al., 2014).HITS-CLIP experiments for all three RBFOX proteins in the mouse brain resulted in the identification of an elaborate target splicing-regulatory network with RBFOX proteins regulating global dynamic spicing changes during mouse brain development.It also highlighted 48 candidate autismsusceptibility genes as direct RBFOX targets, which function in cytoskeleton organization, synaptic transmission, and transcription regulation (Weyn-Vanhentenryck et al., 2014).
The ARE-dependent regulation of mRNAs has been proposed as a NSPC fate decisions’ balancing mechanism.For example, in the adult mouse subventricular zone and cultured NSPCs, it was shown that musashi RNA binding protein 1 (MSI1), an RBP that maintains the proliferation status of NSPCs (Sakakibara et al., 1996), is targeted by ELAVL4 and stabilized in an ARE-dependent manner.This stabilization is considered to be a time-restricted event aiming at prolonging MSI1 activity in proliferating NSPCs preparing to exit the cell cycle even after theMsi1transcriptional inactivation (Ratti et al., 2006).Additionally, the destabilizing RBP AUF1 is highly expressed in proliferating NSPCs, where it promotes the degradation of cell cycle inhibitors,such as the ARE-containing cyclin-dependent kinase inhibitor 1A (Cdkn1A,also known asp21) mRNA.ELAVL2 was shown to antagonize the action of AUF1 by stabilizingCdkn1AmRNA and promoting its translation, favoring the cell cycle exit of progenitor cells.The balance between AUF1 and nELAVL proteins seems to be critical for the proliferation/neuronal differentiation decision of NSPCs (Hambardzumyan et al., 2009).
ZFP36 family members have been extensively studied in immune cells, where they play roles in the regulation of immune and inflammatory responses(reviewed in Makita et al., 2021), however much less is known about their role in neurogenesis.The levels of ZFP36 and ZFP36L1 paralogs have been shown to dramatically drop during neurogenesisin vivo(Dai et al., 2015).ZFP36 downregulation was further proven to be essential for the progression of neuronal differentiationin vitro, and many neuronal specific genes, among which the RBPsNova1,Elavl2,Elavl3, andElavl4, were identified as direct targets of ZFP36 (Dai et al., 2015; Figure 1).Along these lines, we have shown that the downregulation of ZFP36L1 is crucial for the success of the direct neuronal reprogramming of primary mouse astrocytes and identified many neuronal specific genes as its direct targets, among which the RBPsNova1andRbfox1, as well as the cortical neurogenesis TFs thymocyte selection associated high mobility group box (Tox) and RE1 silencing transcription factor(REST) corepressor 2 (Rcor2; Papadimitriou et al., 2023).Recently, ZFP36L1 was identified by CRISPR gene activation screens as a master negative regulator of neuronal differentiation (Black et al., 2020).Single cell RNAseq (scRNA-seq) studies in mouse brains have identifiedZfp36l1as a highly expressed gene in radial precursors of the developing mouse cortex (Yuzwa et al., 2017; Fabra-Beser et al., 2021).Intriguingly, as we will discuss further,Zfp36andZfp36l1have been reported to be targeted by two neurogenic miRNAs, miR-9 (Dai et al., 2015) and miR-124 (Papadimitriou et al., 2023).
The little tailor now demanded the promised reward from the King, but he repented35 his promise, and pondered once more how he could rid himself of the hero
Regulation of neurogenic programs by nonsense mediated decay
Then the first looked round and saw that there was a little hole on his bed, and he said, Who has been getting into my bed? The others came up and each called out, Somebody has been lying in my bed too. But the seventh when he looked at his bed saw little Snow-white, who was lying asleep therein. And he called the others, who came running up, and they cried out with astonishment, and brought their seven little candles and let the light fall on little Snow-white. Oh, heavens! oh, heavens! cried they, what a lovely child! and they were so glad that they did not wake her up, but let her sleep on in the bed. And the seventh dwarf slept with his companions, one hour with each, and so got through the night.
Interplay of RNA Binding Proteins with MicroRNAs in the Regulation of Neurogenesis
Accumulating evidence suggests novel roles for RBPs in the cytoplasm in interfering with miRNA activity, either by hindering miRNA biogenesis or target binding, enabling target mRNA stabilization, or by synergizing with miRNAs in enhancing the downregulation of their targets.miRNAs play crucial roles during brain development and function by targeting a plethora of non-neuronal targets (reviewed in Rajman and Schratt, 2017; Stappert et al., 2018; Shu et al., 2019).Interestingly, many neuronal miRNAs that are gradually upregulated during neurogenesis fine tune the mRNA levels of RBPs orchestrating an intricate post-transcriptional regulatory network during neurogenesis (Figure 1).
Antagonism and synergism of RBPs in miRNA function and biogenesis
PTBP1 has been mostly known for its role in splicing regulation during neurogenesis; however, it has also been reported to interfere with mRNA stability in the cytoplasm.PTBP1 CLIP-seq data from HeLa cells revealed the binding of PTBP1 majorly on intronic regions and 3′UTRs of its target pre-mRNAs (Xue et al., 2009), whereas combination with argonaute RISC catalytic component 2 (AGO2) CLIP-seq data from HeLa showed that PTBP1 regulates mRNA stability through its interplay with miRNAs (Xue et al., 2013).Depending on the relevant position of PTBP1 and miRNA binding sites in the 3′UTR of a transcript, PTBP1 can either compete with miRNA targeting,inducing mRNA stabilization, or –as discussed below – facilitate miRNA action(Xue et al., 2013).One well-studied mRNA that is stabilized by PTBP1 is the component of the REST repressor complex, CTD small phosphatase 1 (Ctdsp1,also known and referred here asScp1; Xue et al., 2013).PTBP1 competes with miR-124 targeting the 3′UTR ofScp1(Xue et al., 2013) and upon downregulation of PTBP1 during neurogenesis,Scp1is efficiently targeted by miR-124 (Visvanathan et al., 2007).Interestingly,Ptbp1is also targeted by miR-124 (Makeyev et al., 2007), revealing a complex interplay between miR-124, PTBP1, and SCP1/REST with an important regulatory role during neuronal differentiation (Xue et al., 2016).
Additionally, the cytoplasmic fraction of RBFOX1 has been shown to stabilize neuronal specific mRNAs by competing with miRNA binding in 3′UTRs of its target mRNAs (Lee et al., 2016; Vuong et al., 2018).More specifically, theCamk2a3′UTR was found to possess 3 binding sites – targeted by miR-26,miR-124, and miR-30 – located very close to the most upstream RBFOX1 binding motif.However, this mechanism is likely only active on a subgroup of cytoplasmic RBFOX1 stabilized target transcripts, as some of them do not contain identifiable miRNA binding sites in their 3′UTRs (Lee et al., 2016).RBFOX1 has been recently reported to stabilizeVamp1mRNA by antagonizing the binding of miR-9.Interestingly, VAMP1 is specifically expressed in inhibitory neurons and this mechanism of VAMP1 regulation by RBFOX1 provides a paradigm of how broadly expressed RBPs in the brain may perform crucial functions in defined neuronal populations (Vuong et al., 2018).
RBPs have been reported to interfere with miRNA biogenesis during neurogenesis, acting at different levels of pri-miRNA maturation (reviewed in Loffreda et al., 2015).PTBP1 has been recently reported to block primiR-124-1 maturation in embryonic stem cells by binding close to the site of pri-miR-124-1 processing and possibly interfering with d
 中國(guó)神經(jīng)再生研究(英文版)2024年9期
中國(guó)神經(jīng)再生研究(英文版)2024年9期
- 中國(guó)神經(jīng)再生研究(英文版)的其它文章
- Does MgSO4 protect the preterm brain? Dissecting its role in the pathophysiology of hypoxic ischemic encephalopathy
- Exosomes derived from microglia overexpressing miR-124-3p alleviate neuronal endoplasmic reticulum stress damage after repetitive mild traumatic brain injury
- On implications of somatostatin in diabetic retinopathy
- Rebuilding insight into the pathophysiology of Alzheimer’s disease through new blood-brain barrier models
- The functions of exosomes targeting astrocytes and astrocyte-derived exosomes targeting other cell types
- Hypothalamic circuits and aging: keeping the circadian clock updated
