Association of MBOAT7 rs641738 polymorphism with hepatocellular carcinoma susceptibility: A systematic review and meta-analysis
Min Lai,Ya-Lu Qin,Qiong-Yu Jin,Wen-Jing Chen,Jia Hu
Abstract BACKGROUND The MBOAT7 rs641738 single-nucleotide polymorphism (SNP) has been proven to influence various liver diseases,but its association with hepatocellular carcinoma (HCC) susceptibility has been debated.To address this discrepancy,we conducted the current systematic review and meta-analysis.AIM To perform a systematic review and meta-analysis on association of MBOAT7 SNP and HCC susceptibility.METHODS We performed a systematic review in PubMed,Web of Science,Scopus,and EMBASE;applied specific inclusion and exclusion criteria;and extracted the data.Meta-analysis was conducted with the meta package in R.Sensitivity and subgroup analyses were also performed.This meta-analysis was registered in PROSPERO (CRD42023458046).RESULTS Eight studies were included in the systematic review,and 12 cohorts from 6 studies were included in the meta-analysis.Our meta-analysis revealed an association between the MBOAT7 SNP and HCC susceptibility in both the dominant [odds ratio (OR): 1.14,95% confidence interval (95%CI): 1.02-1.26,P=0.020] and recessive (OR: 1.21,95%CI: 1.05-1.39,P=0.008) models.Subgroup analysis revealed that stratification of the included patients by geographical origin showed a significant association in Asia (OR: 1.20,95%CI: 1.03-1.39).CONCLUSION This meta-analysis underscores the contribution of the MBOAT7 rs641738 SNP to hepatocarcinogenesis,especially in Asian populations,which warrants further investigation.
Key Words: MBOAT7;Single-nucleotide polymorphisms;Hepatocellular carcinoma;Systematic review;Meta-analysis;Asian populations
INTRODUCTION
Hepatocellular carcinoma (HCC),which ranks as the fourth leading cause of cancer-related mortality worldwide,presents a substantial challenge in the health care landscape[1].Over 80% of HCC cases occur in low-resource and middle-resource countries,particularly in eastern Asia,where medical and social care resources are often constrained[2,3].To address this challenge,early detection,improved management,and careful monitoring of high-risk populations are essential strategies.
Germline DNA single-nucleotide polymorphisms (SNPs) likely represent etiology-specific host factors that determine HCC susceptibility,including SNPs withinPNPLA3(patatin like phospholipase domain containing 3),TM6SF2(transmembrane 6 superfamily 2),andMBOAT7(membrane-bound O-acyltransferase domain-containing 7)[4].The association between thePNPLA3rs738409 SNP and HCC has already been demonstrated by Singalet al[5],who indicated thatPNPLA3is an independent risk factor for HCC.Furthermore,the association between theTM6SF2rs58542926 SNP and HCC has been illustrated by Tanget al[6],who also suggested a significant association of theTM6SF2SNP with HCC risk.
Interestingly,another previously studied SNP is the missense rs641738 C >T variant positioned downstream of theMBOAT7locus.TheMBOAT7rs641738 SNP has been proven to influence histological liver damage in alcoholic liver disease,nonalcoholic fatty liver disease (NAFLD),hepatitis C,and hepatitis B[7].However,its association with HCC has been debated.Thabetet al[8] performed a large case-control study in patients with hepatitis C virus (HCV)-related HCC and found that the role of rs641738 is limited to the early stages of liver disease but not to further progression or occurrence of HCC.Conversely,Donatiet al[9] found that theMBOAT7rs641738 T allele may predispose patients without cirrhosis to HCC.
To address this discrepancy,we conducted the current systematic review and meta-analysis on the association of theMBOAT7SNP and HCC susceptibility,aiming to provide an updated and comprehensive assessment of the evolving evidence in this area.
MATERIALS AND METHODS
Search strategy
We performed a systematic review using the following search strategy in four different databases (PubMed,Web of Science,Scopus,and EMBASE,searched in August 2023): (1): Neoplasms,Hepatic OR Neoplasms,Liver OR Liver Neoplasm OR Neoplasm,Liver OR Hepatic Neoplasms OR Hepatic Neoplasm OR Neoplasm,Hepatic OR Cancer of Liver OR Hepatocellular Cancer OR Cancers,Hepatocellular OR Hepatocellular Cancers OR Hepatic Cancer OR Cancer,Hepatic OR Cancers,Hepatic OR Hepatic Cancers OR Liver Cancer OR Cancer,Liver OR Cancers,Liver OR Liver Cancers OR Cancer of the Liver OR Cancer,Hepatocellular;(2): Membrane bound O-acyltransferase domain-containing 7 protein,human ORMBOAT7protein,human ORMBOAT7OR membrane bound O-acyltransferase domain-containing 7;and (3): (1) AND (2).
Study selection
The inclusion criteria were as follows: (1) Case-control or cohort study evaluating the association betweenMBOAT7rs641738 and HCC risk;and (2) reported odds ratios (ORs) and 95% confidence intervals (95%CIs) and/or allele frequencies and/or genotypes.If duplicate research reports were retrieved,the most comprehensive report was selected to avoid repeated statistics.Studies were excluded if they met one of the following criteria: (1) Review,comment,or conference abstract;and (2) insufficient data to estimate OR and 95%CI.
The following data were extracted from each included study: Author,publication year,country,cohort characteristics,sample size,genotype distribution,allele distribution,minor allele frequency,genotyping method,genetic models,adjustment,and Hardy-Weinberg equilibrium (HWE) data.Two authors independently assessed the studies,and disagreements were resolved through discussion with a third author.
Statistical analysis for meta-analysis
We further conducted a meta-analysis using the cohorts of the included studies.We used theHardyWeinbergpackage (1.7.5) to calculate HWE in the control group of each cohort and performed the meta-analysis using the "metabin","metagen",and "metainf" functions of the meta package (6.5-0) in R (4.3.0).Between-study heterogeneity was examined using theQtest and quantified usingI2.IfI2>50%,heterogeneity was considered significant,and a random-effects model was applied.Otherwise,a common-effects model was used.Sensitivity analysis was carried out by re-estimating pooled ORs and 95%CIs after excluding each eligible study in turn to assess the stability of the pooled results.Publication bias was evaluated using a funnel plot.
This meta-analysis was performed according to the guidelines of the PRISMA[10].The meta-analysis was registered in PROSPERO (CRD42023458046).
RESULTS
Systematic review of association of MBOAT7 rs641738 polymorphism with HCC susceptibility
A total of 153 records were found in four different databases;40 records were found in PubMed,77 in the Web of Science,22 in Scopus,and 14 in EMBASE.After removing 55 duplicates and 24 meeting conferences or records without abstracts,we rigorously reviewed the titles and abstracts of the remaining 74 records.We assessed 24 full-text reports for eligibility and included 8 studies in the systematic review (Figure 1).
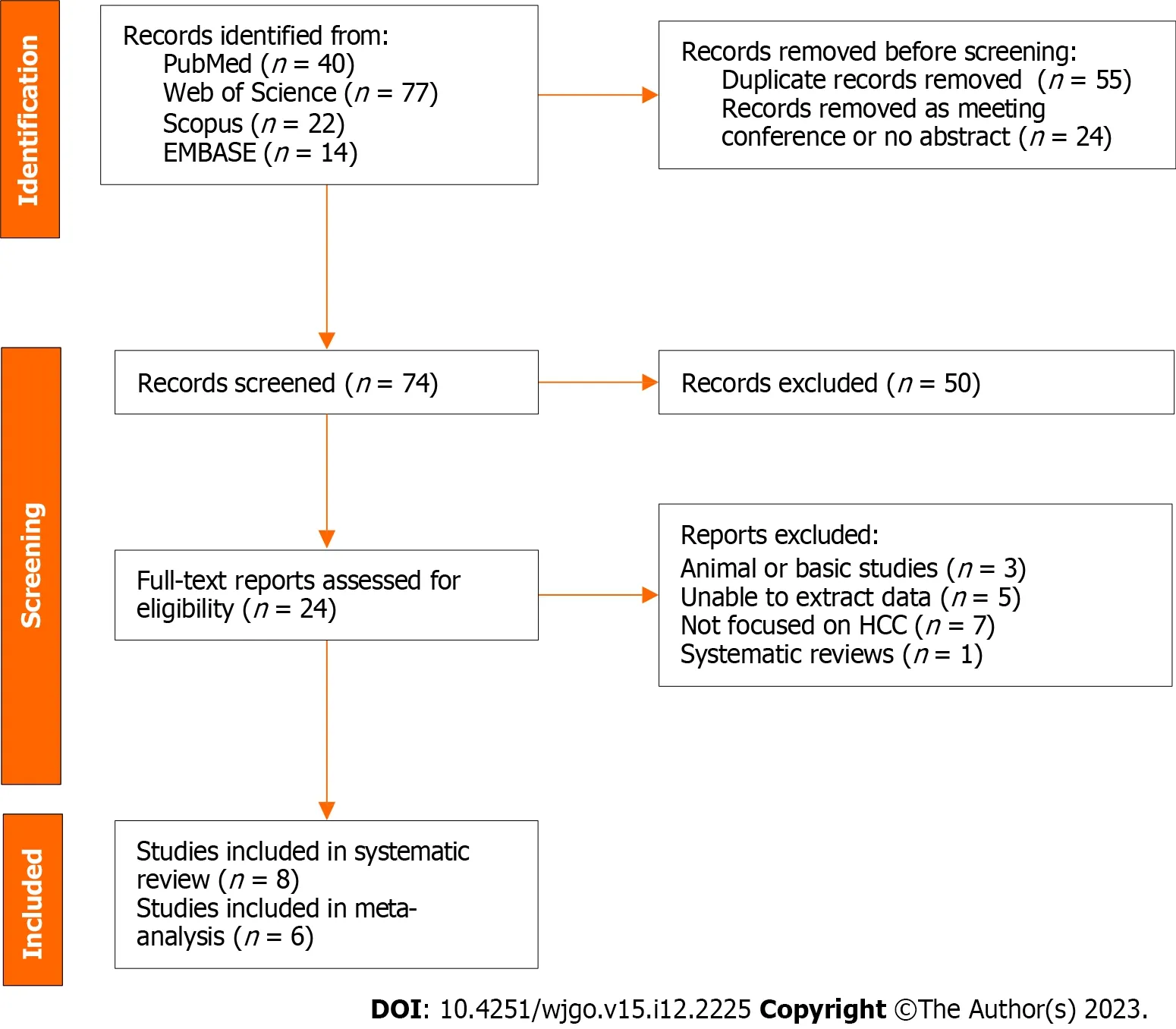
Figure 1 Flow diagram for literature search and study selection.
Detailed information on the included studies is shown in Table 1.Our systematic review encompassed a comprehensive analysis of eight studies,spanning from 2016 to 2022,that explored the connection between theMBOAT7rs641738 polymorphism and HCC.Notably,the majority of these studies (n=6) were conducted outside of Asia.Of these,one study[8] concentrated solely on HCV-related HCC,while another[11] exclusively addressed alcohol-related HCC.Furthermore,two studies focused on HCC arising from NAFLD[12] or metabolic dysfunction-associated fatty liver disease[13],while another two[9,14] investigated a mixed cohort of NAFLD,viral,and alcohol-related HCC.One study[15] encompassed HCC occurring in the context of compensated cirrhosis (HCV or alcohol),and one[16] did not specify the particular characteristics of the included patient cohort.The genetic analysis techniques employed were diverse,with the TaqMan genotyping method being utilized in most studies (n=5),with one study employing the MassARRAY[16] and two[12,13] employing United Kingdom Biobank Axiom array methodologies.A subset of studies (n=4) explicitly delineated the genetic models employed in their analysis.Specifically,two studies[9,11] employed additive models,one[8] adopted a dominant model,and another[16] performed an investigation across dominant,additive,recessive,and allelic models.Conversely,the remaining four studies did not expound upon the genetic models utilized.
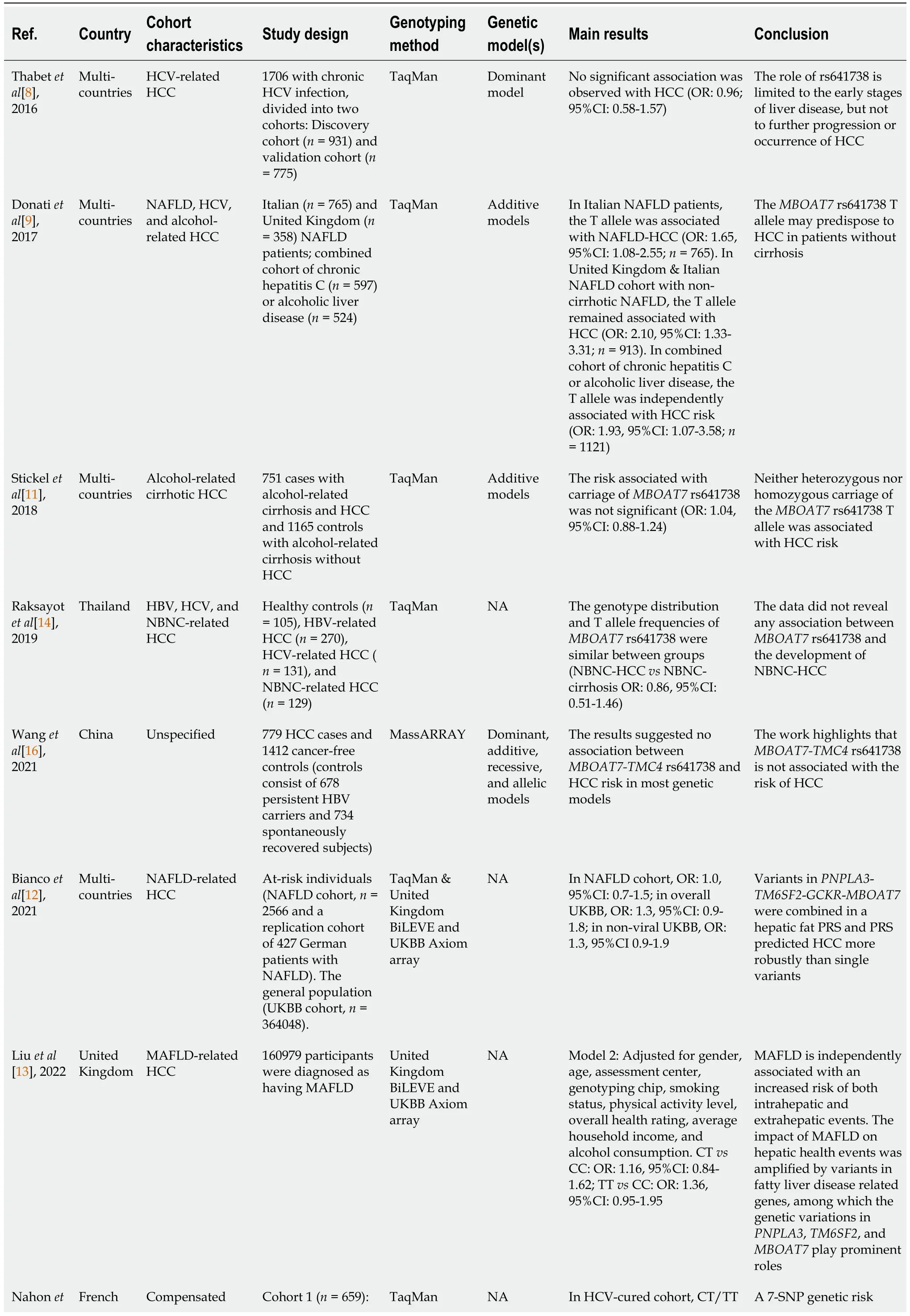
Table 1 Systematic review of MBOAT7 rs641738 and hepatocellular carcinoma susceptibility
Overall,the prevailing trend in the majority of studies (n=7) indicated a lack of significant association between theMBOAT7rs641738 variant and HCC.Among these,Thabetet al[8] examined a cohort of 1706 patients with chronic HCV infection,suggesting that the role of rs641738 was confined to the early stages of liver disease rather than the progression or emergence of HCC.In contrast,Donatiet al[9] conducted an investigation on 765 Italian and 358 United Kingdom NAFLD patients,and they postulated that theMBOAT7rs641738 T allele might predispose individuals to HCC in the absence of cirrhosis.Notably,they discovered that the T allele was autonomously associated with HCC risk in a combined cohort characterized by chronic HCV infection or alcoholic liver disease.To reconcile these divergent findings,both Stickelet al[11] and Raksayotet al[14] pursued separate inquiries,focusing on alcohol-related and virus-related HCC.Intriguingly,both studies arrived at unanimous negative outcomes.In a similar vein,Wanget al[16] undertook a casecontrol study with unspecified cohort attributes,and they predominantly identified negative associations across various genetic models.The remaining three[12,13,15] studies adopted the polygenic risk score (PRS) approach,which encompassed key genes such asPNPLA3,TM6SF2,andMBOAT7.Through PRS analysis,these investigations successfully stratified patients into distinct risk levels for HCC.However,they consistently observed thatMBOAT7'sstandalone predictive capability was comparatively weak.
Meta-analysis of association of MBOAT7 rs641738 polymorphism with HCC susceptibility
In the meta-analysis,12 cohorts from 6 studies were rigorously assessed to elucidate the connection between theMBOAT7rs641738 polymorphism and HCC.To ensure data quality,we excluded the Italian NAFLD-related and HCV-related HCC cohorts from the study by Donatiet al[9] due to HWEPvalues <0.05.Additionally,we excluded the alcohol-related HCC cohort from the same study due to limited sample size (n=12).Employing the "metabin" function,we performed calculations for ORs and 95%CIs,except for the combined Italian and United Kingdom NAFLD-related HCC cohort and the United Kingdom Biobank (nonviral) cohort in the study by Biancoet al[12],in which genotype counts were unspecified.We adopted both dominant and recessive models in the meta-analysis.Cohort characteristics are summarized in Table 2.
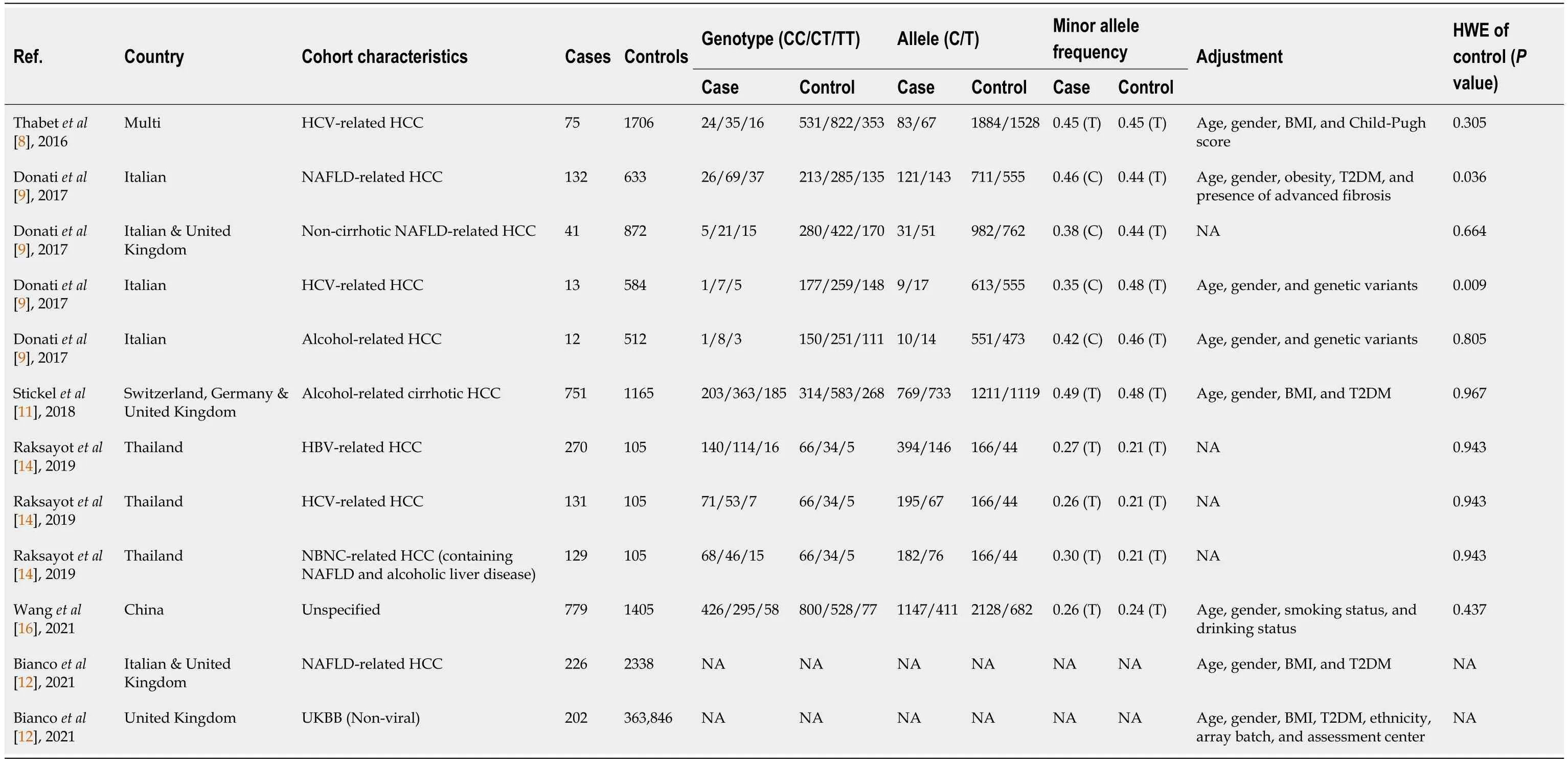
Table 2 Studies included in meta-analysis
Intriguingly,our meta-analysis revealed an association between theMBOAT7rs641738 C >T polymorphism and HCC susceptibility in the dominant model (Figure 2).The pooled OR was 1.14 (95%CI: 1.02-1.26,P=0.020) in the commoneffects model used due to low heterogeneity (I2=33%,P=0.15).Robustness was confirmed in the recessive model (Supplementary Figure 1),with a pooled OR of 1.21 (95%CI: 1.05-1.39,P=0.008).Influential analysisviathe leave-one-out method confirmed the stability of our results (Figure 3).Exclusion of the study by Donatiet al[9] significantly reduced heterogeneity (I2=0%),and the funnel plot indicated this cohort as a source of heterogeneity (Figure 4).Similar outcomes were observed in the recessive model (Supplementary Figures 2 and 3);after omitting Donatiet al[9] or Raksayotet al[14],the heterogeneity was significantly reduced (I2=0%),and thePvalue remained <0.05 in every situation,indicating the robustness of our results.
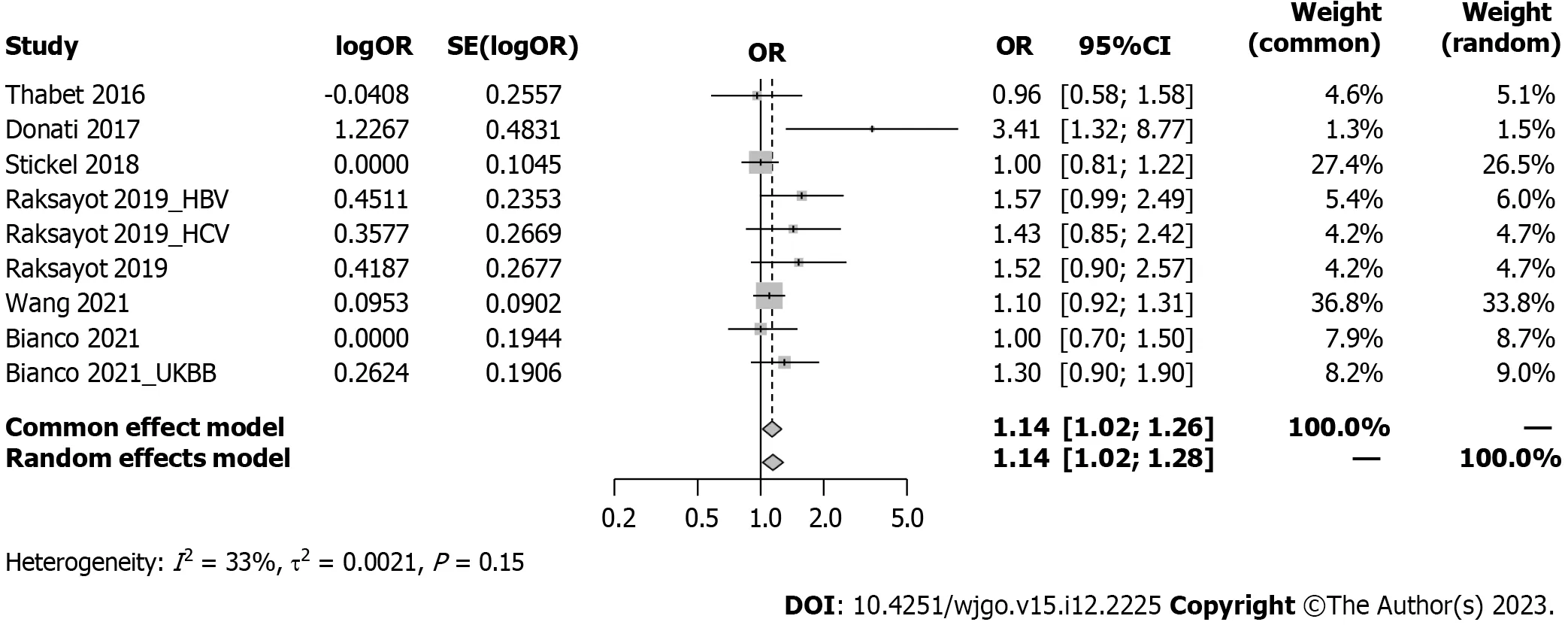
Figure 2 Forest plot of association between MBOAT7 rs641738 and hepatocellular carcinoma risk under the dominant model. OR: Odds ratio;95%CI: 95% confidence interval;HBV: Hepatitis B virus;HCV: Hepatitis C virus;UKBB: United Kingdom Biobank.
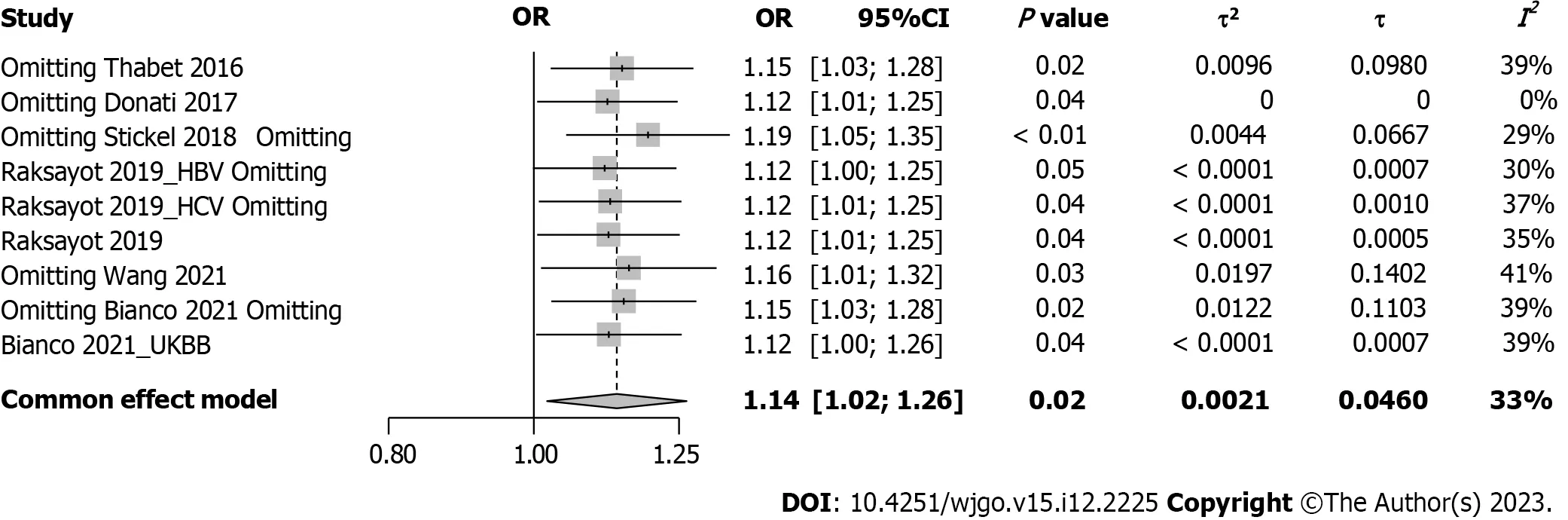
Figure 3 Forest plot of influence analysis under the dominant model. OR: Odds ratio;95%CI: 95% confidence interval;HBV: Hepatitis B virus;HCV:Hepatitis C virus;UKBB: United Kingdom Biobank.
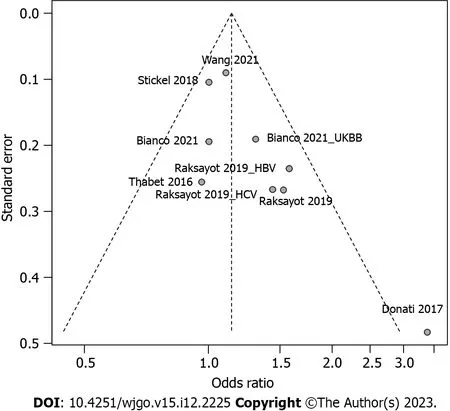
Figure 4 Funnel plot of included studies in meta-analysis under the dominant model. HBV: Hepatitis B virus;HCV: Hepatitis C virus;UKBB: United Kingdom Biobank.
Subgroup analysis considered viral-related and nonviral-related HCC (Figure 5),revealing no significant associations in either the viral (OR: 1.30,95%CI: 0.98-1.73) or nonviral subgroup (OR: 1.09,95%CI: 0.93-1.27).Similar trends were observed in the recessive model (Supplementary Figure 4);in the viral subgroup,the OR was 1.09 (95%CI: 0.69-1.73),while in the nonviral subgroup,the OR was 1.14 (95%CI: 0.96-1.34).Studies were categorized into 5 groups by etiology: HCV,alcohol,NAFLD,hepatitis B virus (HBV),and NAFLD plus alcohol (Figure 6).No significant associations were found in the HCV,HBV,alcohol,or NAFLD subgroup.Notably,in the NAFLD plus alcohol subgroup,the OR was 1.37 (95%CI: 1.01-1.86);however,the same phenomenon was not observed with the recessive model (Supplementary Figure 5).Further stratification by the geographical origin of included patients showed a significant association in Asia (OR: 1.20,95%CI: 1.03-1.39) but not in Europe (Figure 7).In the recessive model,similar results were observed in the Asia subgroup (OR: 1.43,95%CI: 1.06-1.94,Supplementary Figure 6).This observation underscores the need for more investigationwithin the Asian population regardingMBOAT7polymorphisms and HCC susceptibility.
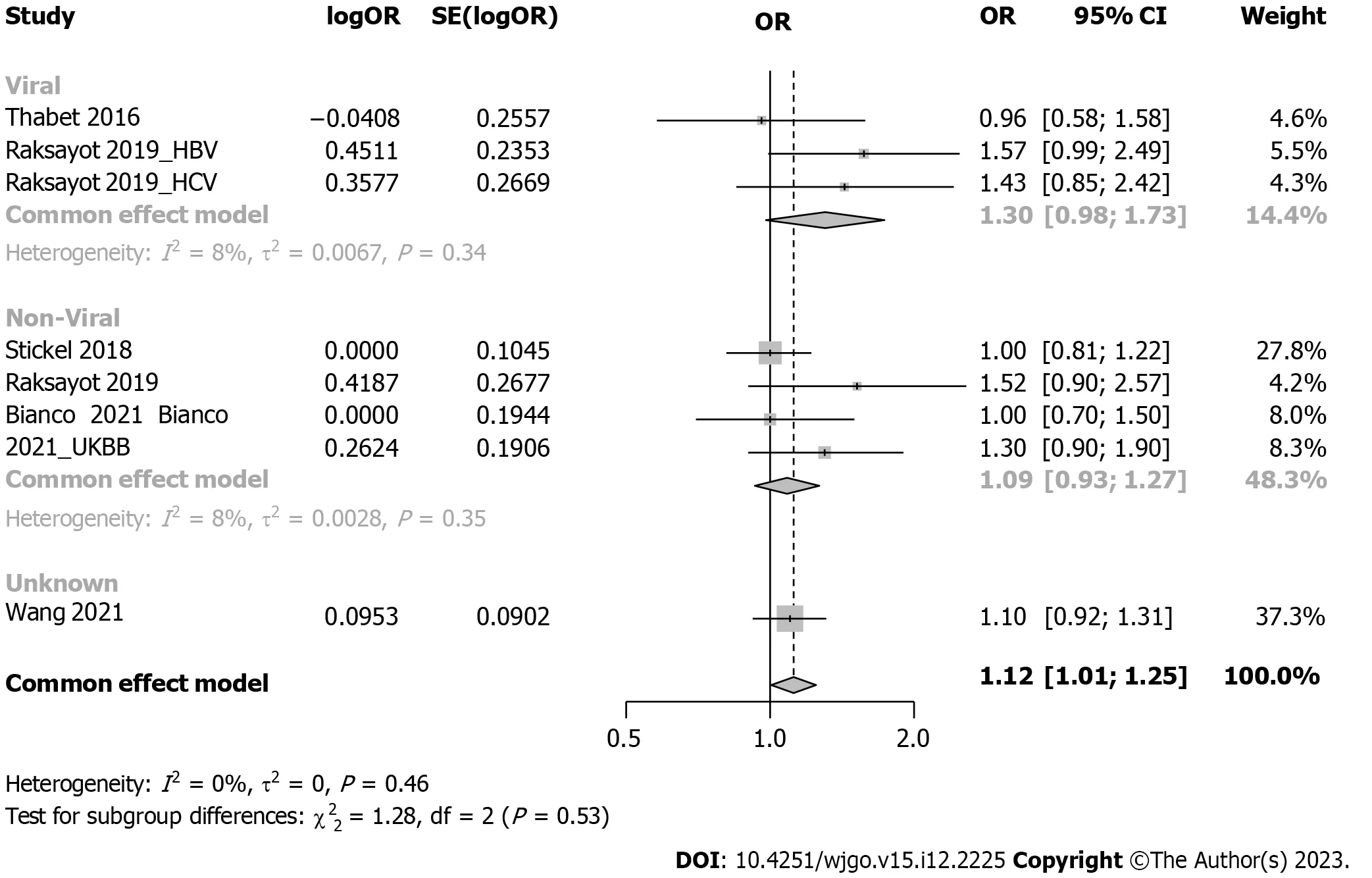
Figure 5 Forest plot of subgroup analysis under the dominant model (stratification by viral or non-viral related hepatocellular carcinoma). OR: Odds ratio;95%CI: 95% confidence interval;HBV: Hepatitis B virus;HCV: Hepatitis C virus;UKBB: United Kingdom Biobank.
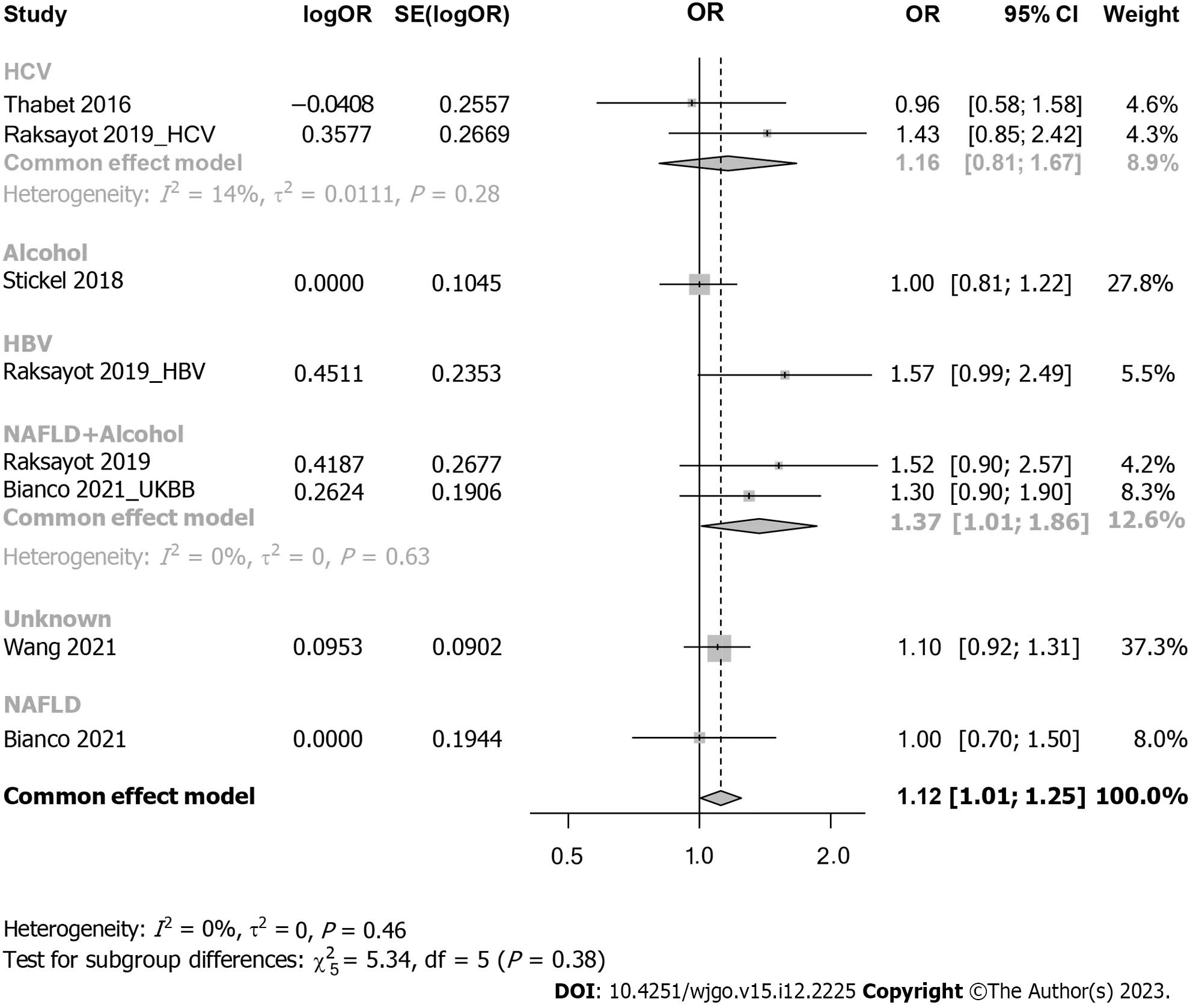
Figure 6 Forest plot of subgroup analysis under the dominant model (stratification by detailed etiology). OR: Odds ratio;95%CI: 95%confidence interval;HBV: Hepatitis B virus;HCV: Hepatitis C virus;UKBB: United Kingdom Biobank.
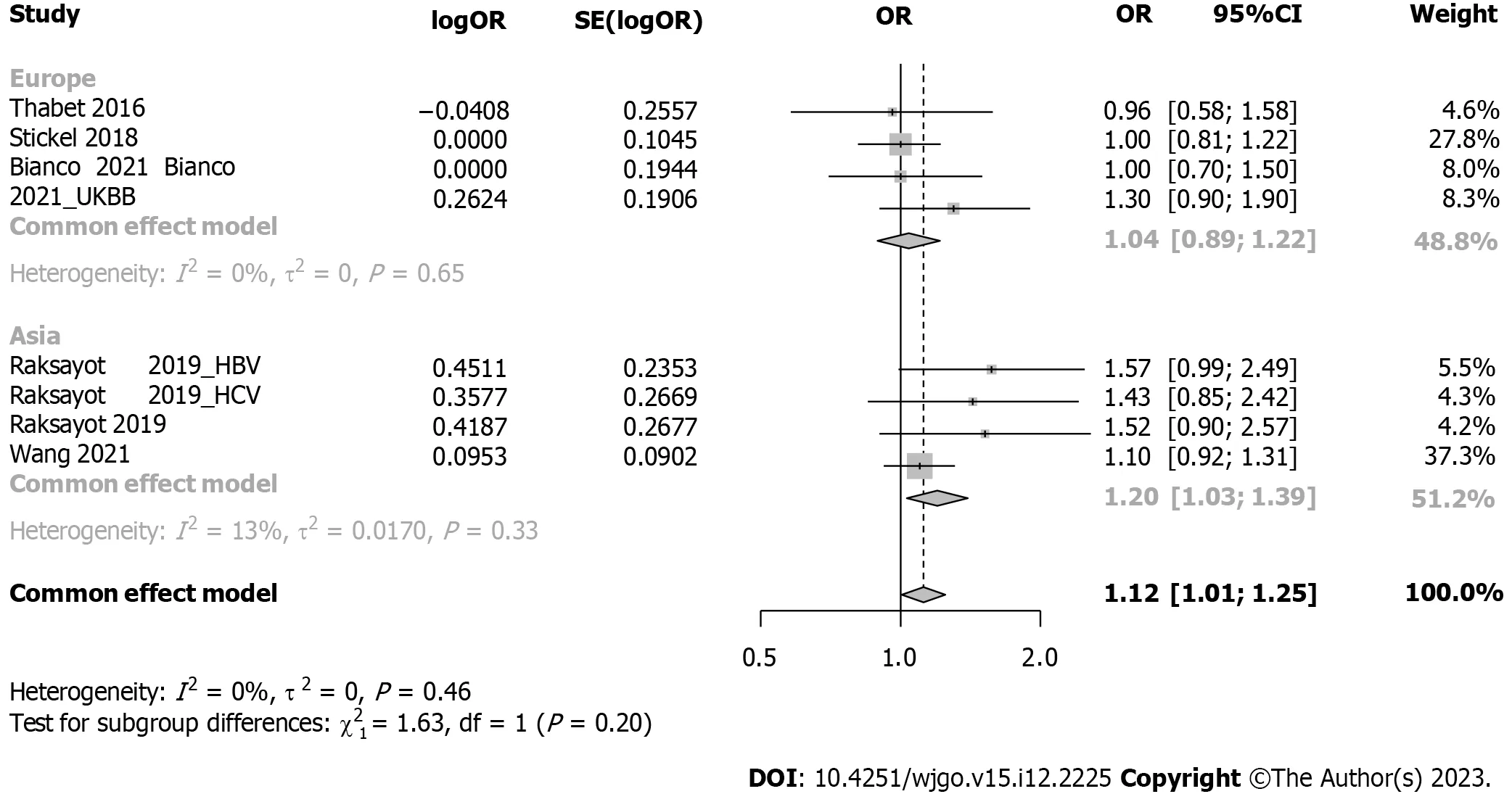
Figure 7 Forest plot of subgroup analysis under the dominant model (stratification by geographical origin). OR: Odds ratio;95%CI: 95%confidence interval;HBV: Hepatitis B virus;HCV: Hepatitis C virus;UKBB: United Kingdom Biobank.
DISCUSSION
In this systematic review and meta-analysis,a total of 2761 HCC cases and 373376 controls were included.Our results suggested that theMBOAT7rs641738 polymorphism was positively associated with HCC susceptibility in both dominant and recessive models.Our subgroup analysis indicated that theMBOAT7rs641738 SNP contributes to hepatocarcinogenesis,especially among Asian populations,warranting further exploration of the underlying mechanisms.
TheMBOAT7rs641738 SNP contributes to hepatocarcinogenesis,which is supported by basic scientific evidence.Longoet al[17] generated a full knockout ofMBOAT7in HepG2 cells (human HCC cell line) and observed an imbalance in mitochondrial dynamics,and the silencing of bothMBOAT7andTM6SF2impaired mitochondrial activity with a shift toward metabolic reprogramming,which led to hepatocarcinogenesis.Moreover,MBOAT7-andTM6SF2-silenced cells exhibited increased cell survival,proliferation,and invasiveness.
Multiple studies have been conducted to investigate the association between the PRS and HCC susceptibility,and most of them reached a positive conclusion.Most studies established PRS models including genes such asPNPLA3,HSD17B13,TM6SF2,MBOAT7,andGCKR.Thriftet al[18] evaluated the association of PRS with HCC in 1644 patients in the United State and found that HCC risk increased by 134% per unit increase in PRS [hazard ratio (HR): 2.30;95%CI: 1.35-3.92].Similarly,Degasperiet al[19] included 509 patients with HCV cirrhosis and found that the PRS was an independent predictor of HCC (HR: 2.30,P=0.04).In contrast,single genetic risk variants were not useful in stratifying HCC risk.Importantly,the PRS method represents a comprehensive assessment of the multiple aspects involved in cancer development.However,the inclusivity of gene sets within the PRS framework remains inconsistent,necessitating further fundamental investigations to refine the approach.
There remain some limitations in this study.First,the included studies in our meta-analysis exhibited diverse etiologies of HCC,which could influence our outcomes.To address this variability,we conducted subgroup analyses and found that the conspicuously positive outcomes might be attributed to studies focused on Asian patients.Additionally,HCC susceptibility could be influenced by patient characteristics,such as age,sex,and smoking habits.Due to the unavailability of the original data of the included studies,this could generate bias in the interpretation of the results.However,the sensitivity analyses confirmed the robustness of our results.
CONCLUSION
In summary,this meta-analysis underscores the contribution of theMBOAT7rs641738 SNP to hepatocarcinogenesis,especially in Asian populations,which warrants further investigation.
ARTICLE HIGHLIGHTS
Research background
TheMBOAT7rs641738 single-nucleotide polymorphism (SNP) has been proven to influence various liver diseases,but its association with hepatocellular carcinoma (HCC) susceptibility has been debated.To address this discrepancy,we conducted the current systematic review and meta-analysis.
Research motivation
Investigating whetherMBOAT7SNP has an association with HCC susceptibility could help identify at-risk population.
Research objectives
We conducted a systematic review and meta-analysis on the association of theMBOAT7SNP and HCC susceptibility,aiming to provide an updated and comprehensive assessment of the evolving evidence in this area.
Research methods
We performed a systematic review in PubMed,Web of Science,Scopus,and EMBASE;applied specific inclusion and exclusion criteria;and extracted the data.Meta-analysis was conducted with themetapackage in R.Sensitivity and subgroup analyses were also performed.
Research results
Eight studies were included in the systematic review,and 12 cohorts from 6 studies were included in the meta-analysis.Our meta-analysis revealed an association between theMBOAT7SNP and HCC susceptibility in both the dominant [odds ratio (OR): 1.14,95% confidence interval (95%CI): 1.02-1.26,P=0.020] and recessive (OR: 1.21,95%CI: 1.05-1.39,P=0.008) models.Subgroup analysis revealed that stratification of the included patients by geographical origin showed a significant association in Asia (OR: 1.20,95%CI: 1.03-1.39).
Research conclusions
This meta-analysis underscores the contribution of theMBOAT7rs641738 SNP to hepatocarcinogenesis,especially in Asian populations,which warrants further investigation.
Research perspectives
Future research should focus on what is the specific molecular biological mechanism ofMBOAT7rs641738 SNP leading to HCC and how to prevent it.
FOOTNOTES
Co-first authors:Min Lai and Ya-Lu Qin.
Author contributions:Lai M and Qin YL conceived,designed,and refined the study protocol;Lai M,Qin YL,Jin QY,Chen WJ,and Hu J were involved in the data collection;Lai M,Qin YL,and Jin QY analyzed the data;Lai M and Qin YL drafted the manuscript;all authors were involved in the critical review of the results and have contributed to,read,and approved the final manuscript.Lai M and Qin YL contributed equally to this work as co-first authors.The reasons for designating Lai M and Qin YL as co-first authors are threefold.First,the research was performed as a collaborative effort,and the designation of co-first authorship accurately reflects the distribution of responsibilities and burdens associated with the time and effort required to complete the study and the resultant paper.This also ensures effective communication and management of post-submission matters,ultimately enhancing the paper's quality and reliability.Second,the overall research team encompassed authors with a variety of expertise and skills from different fields,and the designation of co-first authors best reflects this diversity.This also promotes the most comprehensive and in-depth examination of the research topic,ultimately enriching readers' understanding by offering various expert perspectives.Third,Lai M and Qin YL contributed efforts of equal substance throughout the research process.The choice of these researchers as co-first authors acknowledges and respects this equal contribution,while recognizing the spirit of teamwork and collaboration of this study.In summary,we believe that designating Lai M and Qin YL as co-first authors is fitting for our manuscript as it accurately reflects our team's collaborative spirit,equal contributions,and diversity.
Conflict-of-interest statement:The authors have nothing to disclose.
PRISMA 2009 Checklist statement:The authors have read the PRISMA 2020 Checklist,and the manuscript was prepared and revised according to the PRISMA 2020 Checklist.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers.It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license,which permits others to distribute,remix,adapt,build upon this work non-commercially,and license their derivative works on different terms,provided the original work is properly cited and the use is non-commercial.See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:China
ORCID number:Min Lai 0009-0009-8712-0980.
S-Editor:Lin C
L-Editor:Wang TQ
P-Editor:Zhao S
 World Journal of Gastrointestinal Oncology2023年12期
World Journal of Gastrointestinal Oncology2023年12期
- World Journal of Gastrointestinal Oncology的其它文章
- Dual primary gastric and colorectal cancer: A complex challenge in surgical oncology
- Conversion immunotherapy for deficient mismatch repair locally unresectable colon cancer: A case report
- Prognostic value of T cell immunoglobulin and mucin-domain containing-3 expression in upper gastrointestinal tract tumors: A meta-analysis
- Intensive follow-up vs conventional follow-up for patients with nonmetastatic colorectal cancer treated with curative intent: A metaanalysis
- Paired-related homeobox 1 induces epithelial-mesenchymal transition in oesophageal squamous cancer
- Evaluating the causal relationship between human blood metabolites and gastroesophageal reflux disease
