ldentification of two novel linear epitopes on the p30 protein of African swine fever virus
YU Si-hui ,SHAN Zhao-meng ,YANG Jing-jing ,LlU Yi-ning ,WU Chang-de ,ZHANG Zhen-jiang ,ZHU Yuan-mao,MENG Bo,ZHAN Jia-xing,WEN Xue-xia#,ZHANG Ying#
1 Key Laboratory of Livestock Infectious Diseases of Ministry of Education, College of Animal Science and Veterinary Medicine,Shenyang Agricultural University, Shenyang 110866, P.R.China
2 State Key Laboratory of Veterinary Biotechnology, National High Containment Facilities for Animal Diseases Control and Prevention, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin 150069, P.R.China
African swine fever (ASF) is a hemorrhagic disease caused by African swine fever virus (ASFV),which belongs to theAsfarviridaefamily.ASF has become prevalent in Africa since it was first reported in Kenya in 1921 (Rowlandset al.2008;Costardet al.2009).In 1957,it was introduced to Portugal in Europe,after which ASFV rapidly spread to other European countries and has already been eradicated in several countries except Sardinia (Muret al.2016).In 2007,ASF was introduced to Georgia and has continued its spread to other countries in Europe (Smietankaet al.2016;Kolbasovet al.2018;Gariglianyet al.2019;Lindenet al.2019).In China,an ASF outbreak was first reported on August 3,2018 in a pig farm in Shenyang,Liaoning Province (Zhouet al.2018).Since then,the dramatic spread of ASF throughout China has caused huge economic losses to the swine industry (Jianget al.2021).
ASFV is an enveloped,double-stranded DNA virus.The length of its genome ranges from 170 to 193 kb depending on the isolate,and encodes 150 to 167 proteins which are involved in viral replication,transcription,pathogenesis,and virulence (Chapmanet al.2008,2011;Olesenet al.2018;Wanget al.2020,2022).The p30 protein,a structural protein of ASFV,is encoded by theCP204Lgene (Afonsoet al.1992) and participates in virus internalization,which plays an important role in viral entry (Gomez-Puertaset al.1998).Antibodies against the p30 protein have inhibited the internalization of more than 95% of the virus in macrophages and Vero cells.In addition,a recombinant fusion protein of p30 and p54 can induce neutralizing antibodies (Gomez-Puertaset al.1998;Barderaset al.2001).The p30 protein is expressed during early viral infection stages and can be detected 4 h post-infection in cells.Antibodies against the p30 protein can be detected 8 days after viral infection in pigs (Gimenez-Lirolaet al.2016).Moreover,p30 protein has a strong antigenicity and elicits antibodies with the highest levels during infection (Gimenez-Lirolaet al.2016).The p30 protein is highly conserved among ASFV strains isolated in different countries.Given the properties mentioned above and their critical roles in the replication cycle of ASFV,the development of monoclonal antibodies(mAbs) against p30 could provide valuable tools for the diagnosis and basic research of ASFV.
In this study,the recombinant p30 protein was expressed as a 6× His fusion protein inEscherichia colicells after isopropyl β-D-1-thiogalactopyranoside(IPTG) induction.The protein was purified using affinity chromatography with a Ni-Agarose resin and observed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig.1-A).Its antigenicity was verified using Western blotting analysis with anti-ASFV swine sera (Fig.1-B).The purified p30 proteins were used to subcutaneously immunize 6-week-old female BALB/c mice.The splenocytes from the p30 recombinant proteinimmunized mice were fused with SP2/0 cells.The positive hybridoma cells were screened using p30-specific indirect enzyme-linked immunosorbent assay and then subcloned using limiting dilution analysis.After three subcloning steps,two mAbs against the p30 protein,4F2 and 7F3,were successfully obtained.Isotype determination showed that the 4F2 and 7F3 subclasses were IgG1/κ-type and IgG2b/κ-type respectively (data not shown).Western blotting analysis revealed that 4F2 and 7F3 reacted with the p30 recombinant protein (Appendix A).To further confirm specificity of the two mAbs,the ASFV Pig/HLJ/2018 strain-infected primary porcine alveolar macrophages(PAMs) were subjected to Western blotting and indirect immunofluorescence assay (IFA) (Wenet al.2019;Zhaoet al.2019).Western blotting analysis revealed that 4F2 and 7F3 specifically detected a protein band in the ASFVinfected cells,whereas none were detected in the mockinfected cells (Fig.1-C).In IFA,green fluorescence signals were detected only in the ASFV-infected cells (Fig.1-D),indicating that 4F2 and 7F3 specifically recognized the p30 protein expressed in the virus-infected cells.
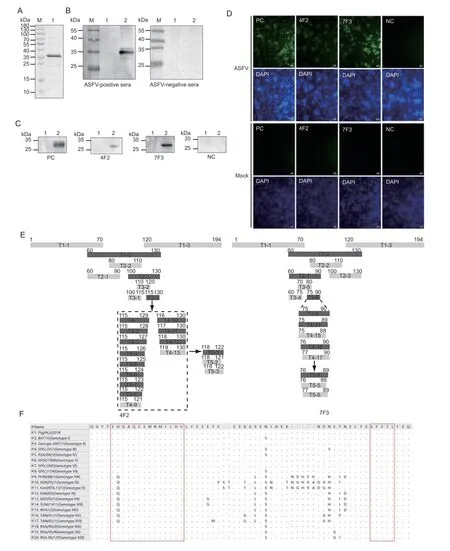
Fig.1 The identification of two novel linear epitopes on the p30 protein of African swine fever virus (ASFV).A,SDS-PAGE analysis of the purified recombinant p30 protein.M,marker;lane 1,purified recombinant p30 protein.B,Western blotting analysis of the purified recombinant p30 protein with anti-ASFV swine sera or ASFV-negative sera.M,marker;lane 1,lysates of pET-30atransformed Escherichia coli with IPTG induction;lane 2,purified recombinant p30 protein.C and D,Western blotting (C) and IFA(D) analyses of the specific reactivity of 4F2 and 7F3 to the p30 protein in the ASFV-infected PAMs.Lane 1,lysates of uninfected PAMs;lane 2,lysates of the ASFV-infected PAMs;PC,positive control (sera from the p30 recombinant protein-immunized mice as primary antibody);NC,negative control (SP2/0 cell supernatants as mock primary antibody).E,schematic diagram showing truncated p30 proteins used for epitope mapping.Rectangles represent truncated p30 proteins,and numbers indicate their amino acid positions.The dark grey rectangles indicate peptides that reacted with 4F2 or 7F3,whereas light grey rectangles represent those that were not recognized by the monoclonal antibodies.F,alignment of the p30 protein sequences from various ASFV strains of different genotypes.The two epitope regions are framed in red.Dots represent residues that are identical to the majority.
The full-length p30 protein was divided into three fragments with 11 overlapping amino acids.The three truncated segments were cloned into the pGEX-6p-1 vector and transformed into BL21 cells for expression.Expression of the GST-fused truncated p30 proteins was confirmed using an anti-GST-tag antibody using Western blotting analysis (data not shown) and further used to verify the reactivity of the truncated p30 proteins to 4F2 or 7F3.The results showed that 4F2 and 7F3 both bound to the T1-2 fragment (Fig.1-E;Appendices B and C).The T1-2 fragment was then divided into 3 overlapping fragments.The T2-3 fragment contained the 4F2 epitope (Fig.1-E;Appendix B),whereas the T2-1 fragment comprised of the 7F3 epitope (Fig.1-E;Appendix C).Subsequently,T2-3 and T2-1 fragments were further divided into three overlapping fragments.Western blotting analysis revealed that 4F2 and 7F3 reacted with fragments T3-3 and T3-6,respectively (Fig.1-E;Appendices B and C).Following this,to precisely define the epitopes recognized by the two mAbs,the T3-3 and T3-6 polypeptides were further individually truncated from their N-and C-termini (Fig.1-E;Appendices B and C).Notably,although T4-15 reacts slightly with 7F3,peptides which had been truncated an amino acid from the C-terminus of T4-15 were not recognized by 7F3 (data not shown).Finally,118SFETL122was shown to be the minimal epitope recognized by 4F2(Fig.1-E;Appendix B),whereas76EHQAQEEWNMILHV89represented that of 7F3 (Fig.1-E;Appendix C).
Currently,there exist 21 epitopes on the p30 protein have been mapped and recorded in the Immune Epitope Database and Analysis Resource (http://www.iedb.org/) (Vitaet al.2019).Among these,15 epitopes of the p30 protein were identified based on the BA71V strain(GenBank accession no.U18466),four from the Georgia 2007/1 strain (GenBank accession no.FR682468),and two on the China/2018/AnhuiXCGQ strain (GenBank accession no.MK128995) (Ivanovet al.2011;Murgiaet al.2019;Petrovanet al.2019;Wuet al.2020;Zhanget al.2021;Zhouet al.2022).Two epitopes from the BA71V strain (111ETNECTSSFETLFEQEPSSE EPKDSKLYMLAQKTVQHIEQYGKAPDFNKV160and91FEEETESSASSESIHEKNDNETNECTSSFETLFEQEPS SE130) and one from the Georgia 2007/1 strain(116TSSFETLFEQ125) contain the newly identified epitope,118SFETL122,which is recognized by 4F2 (Petrovanet al.2019;Wuet al.2020).However,compared with these three epitopes,118SFETL122was more precisely identified and five amino acids shorter.Antigenic regions of the epitope recognized by 7F3 have been characterized in previous studies (Petrovanet al.2019;Wuet al.2020).The amino acid residues 61–110,61–93,and 61–90 of p30 protein from the BA71V strain were identified as epitopes (Petrovanet al.2019;Wuet al.2020).At present,no further fine epitopes have been reported in this region.Our results are the first to show that76EHQAQEEWNMILHV89is an epitope on the p30 protein.
All p30 protein sequences available in GenBank were analyzed to assess the conservation of118SFETL122and76EHQAQEEWNMILHV89epitopes using the MEGA-X Software.In total,569 p30 protein sequences downloaded from the database were aligned.We found that all sequences contained the identical118SFETL122epitope,indicating that mAb-based diagnostic methods are valuable tools for ASFV strain detection.Moreover,73.99% p30 protein sequences harbored the76EHQAQEEWNMILHV89epitope,whereas 26.01%contained a unique amino acid mutation from H to Q at residue 77 (H77Q) (data not shown).The sequence alignment of the p30 protein from representative strains of 19 ASFV genotypes was shown in Fig.1-F,showing an amino acid mutation H77Q within the76EHQAQEEWNMILHV89epitope.Western blotting analysis showed that the H77Q mutation blocked the reactivity of76EHQAQEEWNMILHV89with 7F3 (Appendix C),suggesting that this site is a key amino acid for epitope recognition by 7F3.Sources of the p30 protein sequences harboring the mutated epitope were further analyzed.We found that the p30 protein sequences with the Q residue at the first H site within the epitope were from Africa,including Kenya (Gallardoet al.2011;Bishopet al.2015),Zambia (Simulunduet al.2017),Tanzania,South Africa,Uganda,Zimbabwe,Mozambique,Congo (Bisimwaet al.2021),Burundi,and Malawi (Hakizimanaet al.2021),but not in other parts of the world.Therefore,the combination of 4F2 and 7F3 may be used for the differential detection of such strains.
In summary,we generated two mAbs (4F2 and 7F3) against the ASFV p30 protein and further defined two novel linear B-cell epitopes,118SFETL122and76EHQAQEEWNMILHV89.The two mAbs and identified epitopes have the potential to be used as tools for the diagnosis and basic research of ASFV.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31902258),the Scientific Research Staring Foundation for the Doctors from the Science and Technology Department of Liaoning Province,China(2019-BS-204),and the Major Project of Science and Technology of Liaoning Province (2020JH1/10200003).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
Live viral experiments were performed at the enhanced biosafety level 3 (P3+) facility of the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (Heilongjiang,China) and was approved by the Ministry of Agriculture and Rural Affairs of China (Beijing,China).All animal experiments were conducted in strict accordance with the animal husbandry guidelines of Shenyang Agricultural University (Liaoning,China).Protocols were approved by the Institutional Ethics Committee of the Shenyang Agricultural University(Permit no.2021120201).
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.04.012
 Journal of Integrative Agriculture2023年6期
Journal of Integrative Agriculture2023年6期
- Journal of Integrative Agriculture的其它文章
- Uncertainty aversion and farmers’ innovative seed adoption:Evidence from a field experiment in rural China
- Ensemble learning prediction of soybean yields in China based on meteorological data
- Increasing nitrogen absorption and assimilation ability under mixed NO3– and NH4+ supply is a driver to promote growth of maize seedlings
- Significant reduction of ammonia emissions while increasing crop yields using the 4R nutrient stewardship in an intensive cropping system
- Maize straw application as an interlayer improves organic carbon and total nitrogen concentrations in the soil profile: A four-year experiment in a saline soil
- lnsights into the effects of pulsed antimicrobials on the chicken resistome and microbiota from fecal metagenomes
