Brown planthopper E78 regulates moulting and ovarian development by interacting with E93
ZHENG Shi-wen, JlANG Xiao-juan, MAO Yi-wen, Ll Yan, GAO Han, LlN Xin-da,
1 College of Life Sciences, China Jiliang University, Hangzhou 310018, P.R.China
2 College of Biotechnology and Bioengineering, Zhejiang University of Technology, Hangzhou 310014, P.R.China
Abstract The brown planthopper (Nilaparvata lugens) is the main migratory pest in many rice growing areas in Asia.E78 is a member of the nuclear hormone receptor superfamily which plays an important role in egg development and maternal regulation of early embryogenesis.In this study, brown planthopper E78 (NlE78) was cloned, and the predicted amino acid sequence showed that it contains two conserved domains: NR-LBD and DBD.qRT-PCR showed that the expression of NlE78 is high in the fifth instar nymphs and the ovaries of females.After downregulation of NlE78, the rate of moulting failure (33.2%) increased significantly, and ovarian development was delayed.However, when NlE78 was downregulated together with NlE93, the emergence rate increased significantly (78.79%), and ovarian development was similar to that when NlE78 was downregulated but not delayed.A co-immunoprecipitation experiment showed that NlE78 interacts with NlE93, a crucial downstream transcription factor of the ecdysone signalling pathway.Cellular localization by immunofluorescence revealed that NlE78 and NlE93 are expressed in the nucleus.This study indicates that NlE78 regulates ovarian development and moulting, possibly through its interaction with NlE93.This study is of great significance for the development of new pesticides and control methods based on newly discovered targets.
Keywords: Nilaparvata lugens, ecdysone, E93, E78
1.lntroduction
The brown planthopperNilaparvatalugens(St?l) is one of the main migratory pests in many rice growing areas in Asia (Cheng and Zhu 2006; Linet al.2016a; 2018).The adult brown planthopper has two wing forms.The long-winged brown planthopper tends to migrate from unfavourable habitats, while the short-winged brown planthopper has a high reproductive capacity which enables its adaptation to different environments (Cheng and Zhu 2006; Linet al.2016b).Although chemical control methods are widely used, they often make pests resistant to insecticides (Baoet al.2010).Therefore, it is necessary to explore more targeted, efficient and environmentally-friendly control methods.20-Hydroxyecdysone (20E) is a steroid hormone.In the life cycle of insects, metamorphic development allows them to emerge as adults after several moults.This complicated process relies on the regulation of ecdysone and 20E levels (Yamanakaet al.2013).20E forms a complex with its heterodimeric receptors Ecdysone receptor (EcR) and Ultraspiracle (USP) to regulate the transcription of the early genesBr,E74,E75and?Ftz-F1in the transcription cascade (Evans and Mangelsdorf 2014).Nuclear receptor genes such asEcdysone-inducedprotein78C(E78), Hormone receptor 3 (HR3), Hormone receptor 4 (HR4), Hormone receptorlike 39 (HR39), and E75 are all responsive to ecdysone (Woodardet al.1994; King-Jones and Thummel 2005).
E78is a member of the nuclear hormone receptor superfamily that is induced by 20E in the late stage of insect metamorphosis (Stone and Thummel 1993; Russellet al.1996).The conserved ligand binding and dimerization domain is located in the C-terminal ofDrosophilaE78, and this region contains a heptad repeat sequence of hydrophobic amino acids which can mediate receptor dimerization (Stone and Thummel 1993).It interacts with other members of the superfamily and mediates the effect on chromosome puffs (Stone and Thummel 1993; Russellet al.1996).Thus,DrosophilaE78serves a regulatory role in the ecdysone signalling pathway (Chao and Guild 1986; Huetet al.1993).E78is regulated by the transcripts of early gene clusters and has a high degree of stage specificity (Stone and Thummel 1993).DrosophilaE78is mainly expressed in the pupae and prepupae, and a low transcription level ofE78can be detected in late pupae (Stone and Thummel 1993).E78is expressed throughout the germline during oogenesis (Klieweret al.1992).It plays an important role in regulating stem cell function and maintaining the activity of the follicular germline, controls the viability of cysts, and regulates gamete development and cell apoptosis (Ables and Drummond-Barbosa 2010; Belles and Piulachs 2015; Fingeret al.2021).A recent study found thatE78Ais a central regulator of adult lipid homeostasis and plays an important role in intestinal lipid uptake (Praggastiset al.2021), suggesting thatE78plays a role in lipid metabolism.
The hormone titres appear to peak in the haemolymph of the prepupa, which inhibits metamorphic development under the control of 20E, causing the larvae to undergo moulting (Ashburner 1974; Koelleet al.1991).20E induces the expression of the transcription factorE93, which specifies the onset of adult development, promotes adult development and suppresses larval responses to 20E (Mouet al.2012; Belles and Santos 2014; Liuet al.2015; Lamet al.2022).20E interacts with juvenile hormones to regulate the expression of important regulatory factors, such asE78(Ashburner 1974).Furthermore, 20E cooperates with juvenile hormones to regulate key developmental processes, including yolk production in adult females (Stilwellet al.2003; Sekimotoet al.2007).
Although various functions ofE78have been studied through model systems such asDrosophilamelanogaster, there are still many unknowns.For example, very little is known aboutE78in the brown planthopper.Therefore, this paper investigates the functions ofE78and further explores the possible interactions betweenE78andE93.The results reveal that the interactions betweenE78andE93at the genetic and biochemical levels jointly regulate metamorphosis and development, which provides a theoretical basis for the control of brown planthoppers and therefore provides a new method for pest control.
2.Materials and methods
2.1.lnsect and sampling
The brown planthoppers were raised in our laboratory from insects originally provided by Prof.Zhu Zengrong, Zhejiang University, China.The rice cultivated was IIYou7954 (Zhejiang Academy of Agricultural Sciences, China), which is a newindicahybrid rice combination composed of sterile line II-32A and restorer line Zhehui 7954.Insects were obtained from nymphs at each developmental stage with strictly controlled timing.Forty 1st instar nymphs, 15–30 of each of the 2nd, 3rd, 4th and 5th instar nymphs, and 15 7-day-old brachypterous and macropterous adults were collected for the preparation of qRT-PCR templates.For the refined temporal expression experiment, samples of the 5th instars and females were collected from 0 to 72 h after moulting at 12-h intervals.qRT-PCR templates were prepared from different tissues of the planthoppers one week after emergence.The head, thorax, wing, testis/ovary, midgut and leg were dissected and sampled separately.
2.2.Extraction of total RNA and cDNA synthesis
RNAiso Plus (TRIzol, TaKaRa, Dalian, China) was used to collect the samples, 100–200 μL of TRIzol was added, and the samples were ground.The total RNA was measured with a NANODROP 2000 (Thermo Scientific Inc., USA) after extraction.Reverse transcription was carried out using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Shanghai, China).
KAPA SYBR FAST qPCR Kit (Sigma-Aldrich, Shanghai, China) was used for qRT-PCR.The primers are listed in Appendix A.The following reaction was used: 7.2 μL DEPC-treated water, 10 μL SYBR Green, 0.4 μL upstream and downstream primers, and 2 μL cDNA template.A two-step qRT-PCR amplification program was used as follows: pre-denaturation (1 cycle): 94°C for 3 min; PCR (40 cycles): 94°C for 15 s, and 58°C for 40 s.The data used in the detection were all analysed by the 2–??CTmethod (Schmittgen and Livak 2008).IBMSPSS 20.0 was used for independent samplet-test analysis (P<0.05).
2.3.Cloning and sequence analysis
The primers for the cloning ofNlE78were forward 5′-GGCTTCTTCAGGAGGAGCAT-3′ and reverse 5′-TGTATTTCACTGTAGATCCT-3′ (Sangon Biotech Co., Ltd., Shanghai, China).The newly clonedNlE78was sent to Sangon (China) for sequencing.Editseq Software was used to translate the DNA sequence ofNlE78into its amino acid sequence.E78 homologues were identified and downloaded from NCBI.MegaAlign was used for sequence alignment, and a phylogenetic tree was constructed using MEGA7.
2.4.RNAi
The T7 RNA Synthesis Kit (RiboMAX Large Scale RNA Production Systems-SP6 and T7; Promega, Madison, WI, USA) was used.The primers used for dsRNA synthesis are listed in Appendix B.Primers were used to amplify the target DNA fragments, which were then recovered and purified.Double-stranded RNA was synthesized as previously described (Jinet al.2014).dsRNAs ofNlE93,NlE78andgreenfluorescentprotein(GFP) were synthesized.The concentrations of dsNlE93, dsNlE78and dsGFPwere adjusted to equal proportions when two dsRNAs were used.dsRNA was injected into the brown planthopper using Narishige syringes (MN-151, Narishige, Japan).From the day after injection, we observed and recorded the number of brown planthoppers that moulted and then calculated the survival rate, the proportion of abnormalities and the phenotypes.
2.5.Ovary dissection
Fifth instar brown planthopper nymphs were injected with dsRNAs.The emerging females were collected and mated with males.Three days later, the females were dissected, and their morphology was assessed, recorded and photographed with a microscope (Eclipse 80i, Nikon, Japan).
2.6.Protein–protein interaction
The primers for the constructs are listed in Appendix C.The fragments ofNlE78andNlE93were recovered and purified after restriction enzyme digestion and then subcloned into pXF6F (FLAG tag) and pXF4H (HA tag).All subclones were sequenced for confirmation.
Cell lysates (pH 7.5) were prepared with a lysis buffer composed of 50 mmol L–1Tris-HCl, 75 mmol L–1NaCl, 5 mmol L–1EDTA, NaF (prediluted), Triton X-100, MgCl2, benzonase, and dithiothreitol.SDS buffer was prepared from 5 mL Tris-HCl (pH 6.8), 16 mL SDS, 16 mL glycerol solution, 10 mg bromophenol blue indicator, and 1mL β-mercaptoethanol.
2.7.Co-immunoprecipitation
The cultured cells were primarily 293T cells and HeLa cells.Cells were subcultured in a CO2incubator.When the cell density met the requirements, the medium was changed to NOPS medium (mainly DMEM and foetal calf serum) and an appropriate amount of transfection reagent was added.After transfection, the cells were cultured and harvested, and the lysate was prepared.
The newly prepared cell lysate was centrifuged, and the supernatant was collected.Protein G agarose beads were added and the sample was centrifuged.The supernatant was divided into two parts, and equal volumes of SDS were added to the input group samples.Loading buffer was added to the beads, and the sample was boiled in a water bath.Flag beads were added, and the immunoprecipitate was shaken.Then, the beads were washed at the bottom of the tube, SDS loading buffer was added, and the sample was boiled in a water bath.
The protein sample was subjected to SDS-PAGE and separated by electrophoresis.Then, the samples were transferred to the PVDF membrane at a constant current of 380 mA for 2.5 h.Afterwards, the membrane was blocked with 30 mL skim milk and washed with 1× PBST.The diluted primary antibody (anti-FLAG, Sigma, Shanghai, China; anti-HA, Covance, USA) was added for the incubation (2 h), washed with PBST, and then diluted secondary antibody (anti-mouse/rabbit IgG HRP-linked antibody, CST, USA) was added and the sample was incubated for 1 h.The membrane was washed with PBST and exposed after the addition of HRP substrate.
2.8.Subcellular co-localization
The target genes tagged with HA or Flag were transformed into HeLa cells.Then, the cells were washed with 1× PBS, fixed with PFA/PBS, incubated with Triton/PBS and BSA/PBS, and primary antibody (anti-FLAG, Sigma, Shanghai, China; anti-HA, Covance, USA), washed with 1× PBS, incubated with secondary antibody (Donkey antimouse IgG-Alexa Fluor 488, Donkey anti-rabbit IgG-Alexa Fluor Cy3; Jackson ImmunoResearch, West Grove, PA, USA), and washed with PBS.The nucleus was labelled with DAPI (Sigma, Shanghai, China), and ACA (HCT-0100, Immunovision, USA) was used as a control.
3.Results
3.1.Sequence analysis
The brown planthopperE78was cloned and sequenced, and the amino acid sequences of other homologousE78genes were located and downloaded from NCBI.Sequence analysis revealed that the brown planthopper E78 is composed of a centrally conserved DNA binding domain (DBD), a variable N-terminal domain, a non-conserved hinge, and a C-terminal ligand binding domain (NR-LBD) (Fig.1).The specific binding ligands have been discovered for most of the receptors in the NR-LBD superfamily, but there are also orphan receptors for which no corresponding ligands have been found.Family members include steroid receptors, thyroid hormone receptors (TRαl, FRΔαl and TRΔα2), retinoic acid receptors, receptors for by-products of purine metabolism, glucocorticoid receptors and haem receptors.There is a conserved sequence in the DBD called the P-box (CEGCK) binding site (Fig.1), which is responsible for binding specific DNA target genes at the major groove and forming nonspecific interactions at the minor groove.There is also a zinc finger structure (C4-Type) at the N-terminus of the sequence.Each zinc finger domain contains a group of four cysteine residues, which coordinate a zinc atom and interact with a specific DNA site upstream ofE78to regulate the transcription starting rate.

Fig.1 Alignment of ecdysone-induced protein 78C (E78). NlE78, Nilaparvata lugens (AST48087.1); DmE78, Drosophila melanogaster (NP_001246862.1); LsE78, Laodelphax striatellus (RZF47127.1); CcE78, Cyphomyrmex costatus (XP_018398927.1); DnE78, Dufourea novaeangliae (XP_015431159.1).The blue box represents the DNA-binding domain (DBD), and the red box represents the nuclear receptor-ligand binding domain (NR-LBD).Ligand binding domain (LBD) of nuclear receptor (NR): The nuclear receptor is a transcriptional regulatory domain that is activated by a ligand and regulates the growth and development, reproductive metabolism and other life activities of metazoans.
A phylogenetic tree was constructed to analyse the genetic relationships between brown planthopper E78 and the E78 sequences of various species (Appendix D).The results showed that the brown planthopper E78 is closely related to the E78s of the common bed bug (Cimexlectularius), the small brown planthopper (Laodelphaxstriatellus), the brown marmorated stink bug (Halyomorphahalys), and the rice weevil (Sitophilusoryzae).
3.2.Expression profiles of NlE78
To study the expression profiles ofNlE78in various developmental stages and various tissues, qRT-PCR was performed.The results showed that theNlE78expression levels were higher on the sixth and seventh days of the embryonic stage, approximately 20 times that of the first day (Fig.2).The expression levels of the first to third days of the embryonic stage were relatively low (Fig.2).NlE78was widely expressed at different developmental stages, and its expression was low in eggs and adults (Fig.2).The expression in the nymphs was significantly higher than that of adults, especially 5th instar nymphs, which was approximately 9–10 times that of adults.The 2nd and 3rd instar nymphs had higher expression levels, and the 1st and 4th instar nymphs had lower expression levels (Fig.2).
The expression ofNlE78reached the highest level in the testis of the long-winged male, which was at least 3–9 times those of the other tissues (Fig.2).The expression in the ovary of the long-winged females was approximately 12 times that of the midgut, followed by the leg, brain, thorax, forewings and midgut (Fig.2).In the short-winged females, the expression in the brain was 145 times that of the leg, followed by the ovary and midgut.In short-winged males, the highest expression (forewing) was approximately 9 times that of the thorax (Fig.2).The expression ofNlE78in the 5th instar nymph increased until 60 h after moulting (Fig.2-D).However, the expression ofNlE78was relatively stable after eclosion in females (Fig.2-E).
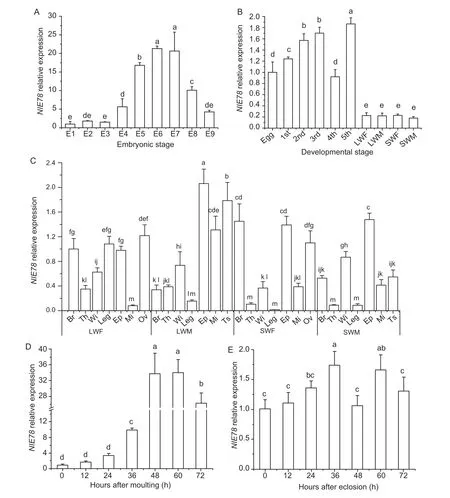
Fig.2 Spatial and temporal expression profiles of NlE78.A, expression of NlE78 at different embryonic stages.E1–E9 indicate embryos 1–9 days after eggs laid, respectively.B, expression of NlE78 at different developmental stages.1st, 2nd, 3rd, 4th, and 5th, indicate 1st, 2nd, 3rd, 4th and 5th instar nymph, respectively; LWF, long-winged female; LWM, long-winged male; SWF, short-winged female; SWM, short-winged male.C, expression of NlE78 in different tissues.Br, brain; Th, thorax; Wi, forewing; Leg, leg; Ep, epidermis; Mi, midgut; Ov, ovary; Ts, testis.D, expression of NlE78 in the 5th instar nymph at 0–72 h after moulting.E, expression of NlE78 in females at 0–72 h after eclosion.One-way analysis of variance (Duncan’s for multiple comparisons method) was used.The letters indicate whether there is a significant difference, the same letter indicates no significant difference, different letters indicate a significant difference between the two (P<0.05), and the farther apart the letters are alphabetically, the greater the difference between the two.Error bars indicate SD (n=3).
3.3.Downregulation of NlE78 affected development and eclosion
E78is involved in the regulation of various pathways, such as follicular development, early embryonic development and extracellular signal transduction (Ables and Drummond-Barbosa 2010; Belles and Piulachs 2015; Fingeret al.2021).When the 5th instar nymphs were injected with dsNlE78and dsNlE93, alone or in combination, the moulting of the brown planthopper was affected (Fig.3-B–D).The epidermis on the back became cracked, the body stretched, and the nymphs could not emerge (Fig.3-B–D).We then recorded and counted the number of surviving and emerged brown planthoppers (Fig.3-E–G).The results showed that the eclosion rate of the 5th instar nymphs after RNAi was lower than that of the control injected with dsGFP(Fig.3-E–G).WhenNlE78was downregulated alone, the emergence rate was 66.8%; whenNlE93was downregulated alone, the emergence rate was 70.2%; and whenNlE93andNlE78were downregulated together, the emergence rate increased to 78.8%, suggesting a possible interaction between the two genes (Fig.3-G).
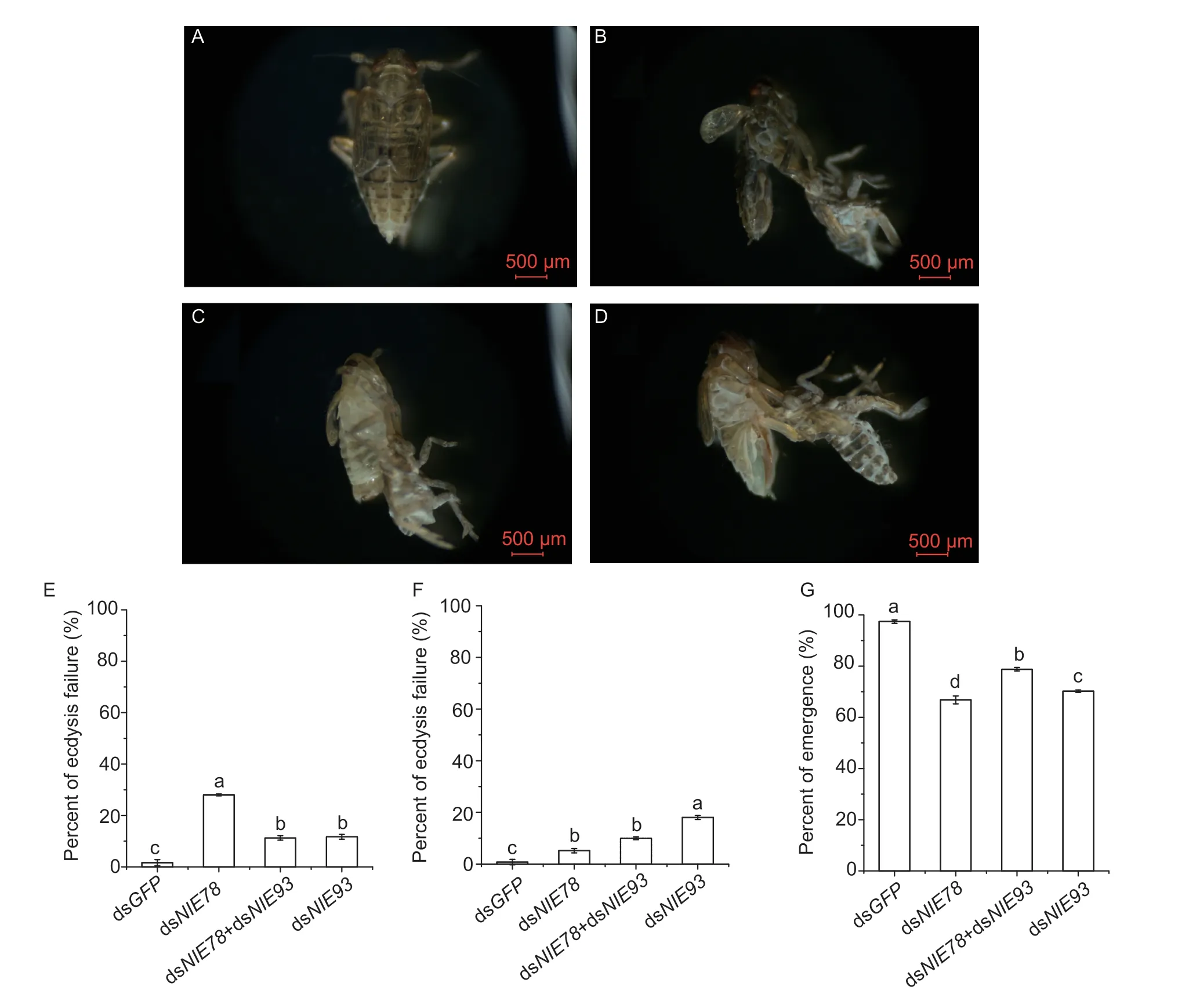
Fig.3 Effect of downregulating NlE78 and NlE93 in 5th instar nymphs. A–D, images of brown planthopper injected with dsGFP, dsNlE93, dsNlE78, and dsNlE78+dsNlE93, respectively.E, percentage of ecdysis failure for 5th instar nymphs.F, percentage of ecdysis failure.G, percentage of emergence.One-way analysis of variance (Duncan’s test for multiple comparisons) was used.Error bars indicate SD (n=3).Different letters indicate significant differences (P<0.05).
3.4.Downregulation of NlE78 delayed ovarian development
The downregulation ofNlE78andNlE93, alone or in combination, affected ovarian development.After downregulating the expression ofNlE78, the ovarian tubules did not develop, there were no egg chambers, and the ovaries were small (Fig.4-C).After downregulating the expression ofNlE93, ovarian development was arrested (Fig.4-B).However, whenNlE93was downregulated together withNlE78, ovarian development was slower but not arrested (Fig.4-D).
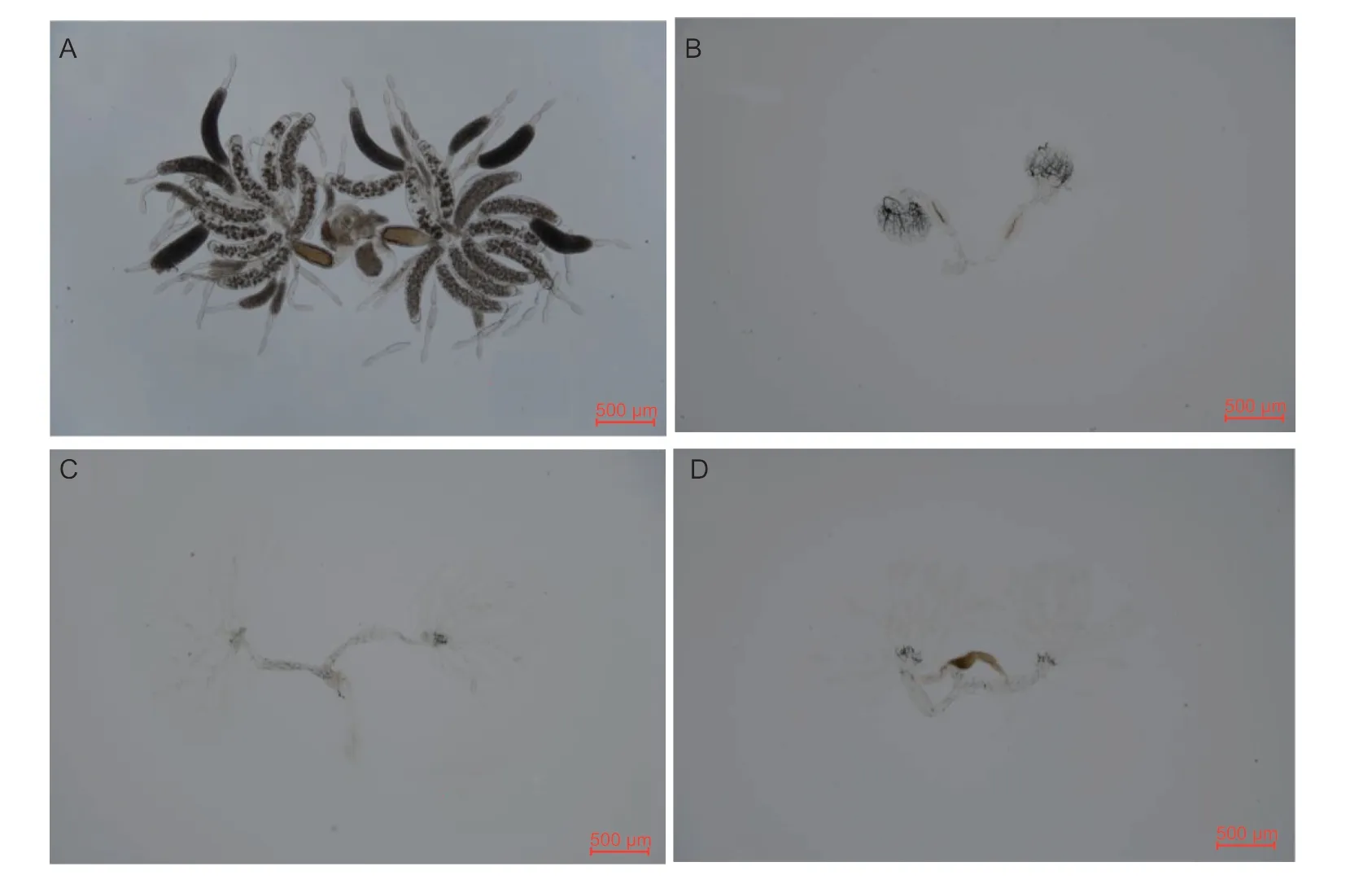
Fig.4 Ovarian development after downregulating NlE78, NlE93 or both.A, dsGFP.B, dsNlE93.C, dsNlE78.D, dsNlE78+dsNlE93.
3.5.Downregulation of NlE78 affected the expression of ecdysone-related genes
To test whether the expression of ecdysone-related genes was affected by downregulatingNlE78, we downregulatedNlE78by RNAi.After RNAi was performed on 3rd, 4th and 5th instar nymphs, qRT-PCR was used to evaluate the RNAi efficiency.The results showed that after RNAi was carried out on nymphs of different instars, the expression of the target gene was significantly reduced, which indicated that the RNAi was effective (Appendix E).
After 3rd, 4th and 5th instar nymphs were injected with dsNlE78and dsGFP(control), the expression levels ofNlECR,NlUspA,NlUspB,NlE93andNlBr(ecdysone signalling pathway) were measured by qRT-PCR.After downregulatingNlE78at the 3rd instar nymph stage, the relative expression ofNlE93decreased (Fig.5-A).After downregulatingNlE78, the expression levels ofNlECR,NlUspBandNlBrincreased (Fig.5-A).After downregulatingNlE78at 4th instar nymph stage, the relative expression ofNlE93andNlBrdecreased (Fig.5-B).After downregulatingNlE78at 5th instar nymph stage, the relative expression levels ofNlECR,NlUspA,NlE93andNlBrdecreased significantly (Fig.5-C).We also measured the expression ofNlE78after downregulating the hormone-related genes.The results showed that the expression ofNlE78increased significantly after downregulatingNlE93,NlUspA,NlUspBandNlKr-h1(Fig.5-D).In summary, the expression ofNlE93was reduced in all three nymphal stages we tested, indicating thatNlE78promotes the transcription ofNlE93directly or indirectly by changing the 20E titre (Fig.5).
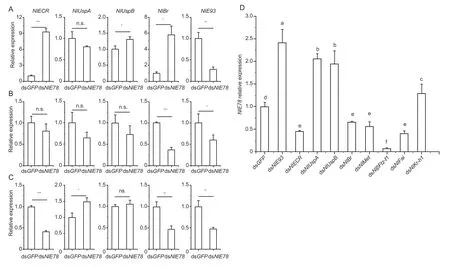
Fig.5 Expression of ecdysone-related genes and NlE78 after RNAi.A–C, downregulating NlE78 at 3rd, 4th and 5th instar nymph stage, respectively.D, expression of NlE78 after downregulating ecdysone-related genes.Samples were collected three days after dsRNA injection.Ecdysonerelated genes are NlECR, NlUspA, NlUspB, NlBr and NlE93.Student’s t-test was used.Error bars indicate SD (n=3).*, P<0.05; **, P<0.01; ***, P<0.001; ns, no significance.
3.6.NlE78 interacts with NlE93 and they are colocalized in the nucleus
To further explore the possible interactions between NlE78 and a crucial transcription factor of the ecdysone signalling pathway, NlE93, we used co-immunoprecipitation (Co-IP) to test the possible protein–protein interactions.We subcloned NlE78 and NlE93 into the Co-IP plasmid (containing Flag or HA).The results showed that NlE78 and NlE93 can be co-immunoprecipitated (Fig.6-A), indicating that NlE78 and NlE93 interact at the protein level.
We used immunofluorescence to determine the subcellular locations of NlE93 and NlE78.The results showed that these two proteins are colocalized in the nucleus (Fig.6-B), suggesting that NlE78 and NlE93 may serve transcriptional regulatory functions.
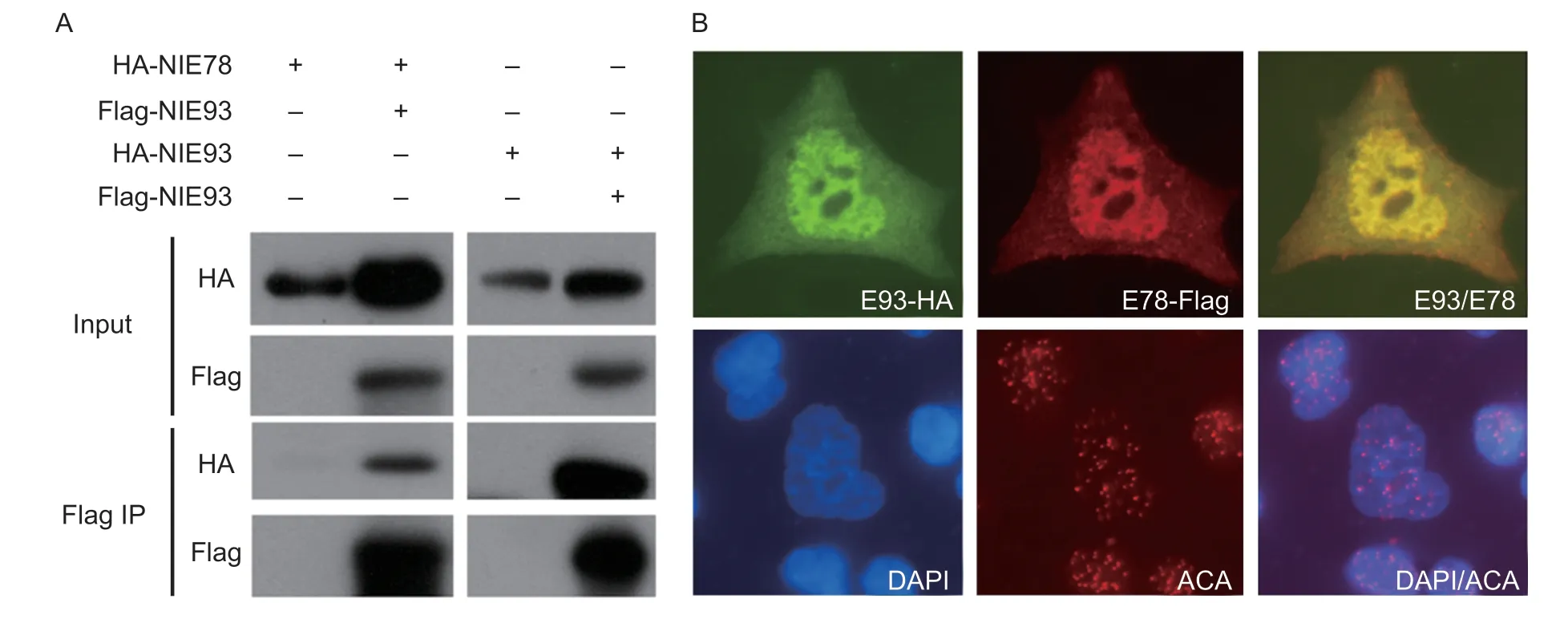
Fig.6 NlE78 interacts with NlE93 and colocalizes in the nucleus.A, coexpression of HA-NlE78 and Flag-NlE93, Flag-NlE78 and HA-NlE93 in 293T (HEK 293) cells.Input, control; Flag IP, immunoprecipitated by anti-FLAG antibody.B, subcellular localization of NlE78 and NlE93, coexpression of HA-NlE93 and Flag-NlE78 in HEK 293 cells.Anti-HA antibody was used to show the subcellular localization of NlE93 (green) and NlE78 (red).DAPI (blue) was used to show the nucleus, and anti-centromere antibody (ACA) was used as an internal control (red dots).
4.Discussion
This study found that the predicted amino acid sequence ofN.lugensE78 is highly conserved with the homologues of other species (Fig.1).qRT-PCR showed that E78 was expressed in 5th instar nymphs and females (Fig.2).We speculated that this gene has a regulatory effect on the metamorphosis and ovarian development of brown planthoppers.We found that whenNlE78was downregulated, the expression of ecdysone-related genes changed consistently, especially the expression ofE93, which implies thatNlE93and the late transcription factorNlE78may be correlated.Further experiments on the downregulation ofNlE78andNlE93suggested their potential genetic interaction.WhenNlE78was downregulated together withNlE93, the emergence rate increased significantly, and ovarian development was similar to that whenNlE78was downregulated but not arrested, which was similar to the downregulation ofNlE93(Figs.3 and 4).Therefore, we speculate that downregulating the expression ofN.lugensE78alone, or downregulating the expression ofNlE78andNlE93at the same time, will also affect the expression of ecdysonerelated genes through genetic interactions betweenNlE78andNlE93, which then further affects reproduction, survival and ovarian development.
Moreover, our co-immunoprecipitation (Co-IP) result (Fig.6) supported the interaction between NlE78 and NlE93, i.e., we detected an interaction between NlE78 and NlE93 at the protein level (Fig.6).Furthermore, a subcellular localization experiment showed that NlE78 and NlE93 are colocalized in the nucleus.The ovary and eclosion phenotypes after RNAi are reminiscent of previous studies in the model organismDrosophilamelanogaster, in which E78 is an ecdysone-responsive gene (Stone and Thummel 1993) that controls the stem cell niche formation of ovarian germline stem cells (Ableset al.2015), while E93 controls the metamorphosis of insects and directly regulates the formation of adult tissues and organs (Belles and Santos 2014; Urenaet al.2014; Maoet al.2019).Thus, we speculated that NlE78 and NlE93 interact at the protein level, regulate the transcription of ecdysone-related genes, and ultimately regulate ovarian development and moulting in brown planthoppers.
5.Conclusion
The brown planthopperE78has a regulatory role in ovarian development and moulting, possibly through interaction withE93.Thus, targeting the interactions between the key transcription factors known in the ecdysone signalling pathway could be of great significance for developing novel measures to prevent and control this northern rice pest.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (32172390, 1672023 and 31741107) and the Natural Science Foundation of Zhejiang Province, China (LZ20C140002).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2022.08.106
 Journal of Integrative Agriculture2023年5期
Journal of Integrative Agriculture2023年5期
- Journal of Integrative Agriculture的其它文章
- Herbicidal activity and biochemical characteristics of the botanical drupacine against Amaranthus retroflexus L.
- Developing a duplex ARMS-qPCR method to differentiate genotype l and ll African swine fever viruses based on their B646L genes
- The effects of maltodextrin/starch in soy protein isolate–wheat gluten on the thermal stability of high-moisture extrudates
- Elucidation of the structure, antioxidant, and interfacial properties of flaxseed proteins tailored by microwave treatment
- Effects of planting patterns plastic film mulching on soil temperature, moisture, functional bacteria and yield of winter wheat in the Loess Plateau of China
- lnversion tillage with straw incorporation affects the patterns of soil microbial co-occurrence and multi-nutrient cycling in a Hapli-Udic Cambisol
