mgr-mir-9 implicates Meloidogyne graminicola infection in rice by targeting the effector MgPDI
TIAN Zhong-ling, ZHOU Jia-yan, ZHENG Jing-wu, HAN Shao-jie,
1 Key Laboratory of Pollution Exposure and Health Intervention of Zhejiang Province, Interdisciplinary Research Academy (IRA), Zhejiang Shuren University, Hangzhou 310015, P.R.China
2 Laboratory of Plant Nematology, Institute of Biotechnology, College of Agriculture & Biotechnology, Zhejiang University, Hangzhou 310058, P.R.China
3 Ministry of Agriculture Key Lab of Molecular Biology of Crop Pathogens and Insect Pests, Zhejiang University, Hangzhou 310058, P.R.China
Abstract MicroRNAs (miRNAs), a class of small non-coding RNAs, are crucial endogenous gene regulators in a range of animals, including plant-parasitic nematodes.Meloidogyne graminicola is an obligate sedentary endoparasite of rice and causes significant yield losses.A number of studies focused on the roles of M.graminicola effectors during the parasitic process; however, how nematode miRNAs regulate its effectors needs elucidating.In this research, we analyzed a cluster of M.graminicola miRNAs obtained at the second-stage juveniles (J2s) stage that are closely linked to the regulation of M.graminicola effectors.There are 49 767 105 total clean reads obtained from three libraries.A total of 233 known miRNAs and 21 novel miRNAs were identified.Among the known miRNAs, mgr-lin-4, mgr-mir-1, mgr-mir-100, mgrmir-86, mgr-mir-279, mgr-mir-87, mgr-mir-71, mgr-mir-9, mgr-mir-50, mgr-mir-72, and mgr-mir-34 are the most abundant 11 miRNAs families.Moreover, the expression levels of selected miRNAs were validated by real-time quantitative PCR.We hypothesized that these miRNAs might regulate the expression of secreted effectors during the J2s stage to facilitate its infection.Consistent with this, we found that mgr-mir-9 targets MgPDI, an important M.graminicola effector mRNA.In addition to that, J2s treated with mgr-mir-9 mimics showed down-regulation of MgPDI expression and reduced reproductive ability, alluding mgr-mir-9 is involved in nematode infection.These results provide novel insight into the regulatory functions of M.graminicola miRNAs during the infection and identify miRNAs and their effector targets as potential key management targets to limit parasite survival during the early stages of infection.
Keywords: microRNA function, Meloidogyne graminicola, deep sequencing, Dual-Luciferase Reporter Assay System, protein disulfide isomerase
1.Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that serve as endogenous gene regulators.Since the discovery of miRNA, lin-4, in the free-living nematodeCaenorhabditiselegans(Leeet al.1993), thousands of miRNAs have been identified in various organisms including humans, insects, nematodes, plants, and viruses.It has been demonstrated that miRNAs play a critical role in many biological processes, such as reproduction, development, organ differentiation, response to a stressor, and pathogenesis of diseases (Carrington and Ambros 2003; Ambros 2004; Bartel 2004; Chen 2005; Zhanget al.2007; Zhang and Wang 2015; Gebert and MacRae 2019).For example, microRNA-9 (miR-9), one of the most highly expressed microRNAs in the developing and adult vertebrate brain, play a prominent role in balancing proliferation in embryonic neural progenitor populations (Coolenet al.2013).miRNAs act as posttranscriptional regulators of their messenger RNA (mRNA) targetsviabinding to their mRNA targets (Schnall-Levinet al.2010; Zisouliset al.2010).The miRNA recognition sequence mainly at the 3′ untranslated region (UTR) of the target mRNA.However, some miRNA could also pair to its responsive element in 5′ UTR sequences as well as in coding regions of mRNAs.In plants, perfect complementarity between miRNA and target mRNA is required for the decay of target transcripts.In animals, miRNAs disrupt their target mRNA through imperfect pairing with complementary sites (Filipowiczet al.2008).
Root-knot nematodes (RKNs,Meloidogynespp.) are the major plant pathogens worldwide (Trudgill and Blok 2001; Castagnone-Serenoet al.2013).In particular,M.graminicolacauses significant yield reductions and is listed as a quarantine pest in most rice-growing countries (Bridge and Page 1982; Pokharelet al.2010; Kyndtet al.2014).Meloidogynegraminicolais an obligate sedentary endoparasite that completes its lifecycle within the rice roots.The pre-parasitic second-stage juveniles (J2s) penetrate the epidermal root tip by secreting enzymes and continuous stylet thrusting.After establishing the feeding site known as giant cells that serve as a source of nutrition,M.graminicolaJ2s undergo physical development and transform into adult stages.The male moves out from the root while the females become sedentary and remain embedded within the root tissue (Kyndtet al.2014).Unlike other RKNs,M.graminicolacan be completely buried under the epidermis of host roots, and the oocysts are not exposed (Kyndtet al.2014).
A number of RKN–host interaction studies focus on the functions of effectors produced in the nematode esophageal glands.The RKN effectors play a crucial role in suppressing basal host defense responses during compatible interactions with their host (Hewezi and Baum 2013; Quentinet al.2013).For example, theM.graminicolaeffectors MgGPP and MgMO237 promote the infection process by inhibiting cell death and suppressing rice basal immunity, respectively (Chenet al.2017, 2018).Our previous studies also showed that twoM.graminicolaeffectors, protein disulfide isomerases (PDIs) could induce plant cell death and contribute to nematode parasitism (Tianet al.2019, 2020).Investigating the function and regulation of effector genes is vital for devising nematicides and nematode resistance breeding (Ferriset al.2012).However, little is known about the mechanisms on spatio–temporal regulation ofM.graminicolaeffector expression.
As integral regulators, the plant miRNAs were reported to play significant roles in plant–nematode interaction (Cabreraet al.2016; Kauret al.2017; Tian Bet al.2017; Panet al.2019).In rice, host small and long noncoding RNAs were identified and characterized in the interaction between rice andM.graminicola(Verstraetenet al.2021).In plant-parasitic nematodes, the proposed functions of miRNA expressed in different lifecycle stages are revealed by small RNA sequencing.Recent studies onM.incognita, another species of the RKNs, suggested their miRNAs could modulate certain target gene levels during nematode parasitism (Wanget al.2015; Subramanianet al.2016; Zhanget al.2016).However, the role ofM.graminicolamiRNAs in regulating effector genes has not been reported.
In this study, we aimed to identify and investigate the role ofM.graminicolamiRNAs in nematode parasitism.We sequenced the miRNAs ofM.graminicolaJ2s and identified 233 known miRNAs and 21 novel miRNAs.AmongM.graminicolamiRNAs, mgr-mir-9 was validated to regulate the expression of the effector gene,MgPDI, and facilitate infection.We anticipate our findings will contribute to the in-depth understanding of miRNA functions within the parasite.
2.Materials and methods
2.1.Meloidogyne graminicola culture and main- tenance
The rice line used in this work wasOryzasativacv.Nipponbare, seeds were germinated on wet filter paper for 8 d at 28°C and transferred to potting soil in a growth chamber.The growth chamber conditions are as follows: 16 h/8 h light/dark photoperiod at 28°C/26°C and relative humidity of 75%.TheM.graminicolaisolates ZJJH was maintained on rice in potting soil in a growth chamber (Huanget al.2016).
2.2.Preparation of specimens
Approximately 5 000 pre-parasitic J2s were collected as described by Huanget al.(2016) and Haegemanet al.(2016).The samples were collected quickly.Then, the samples were snap-frozen in liquid nitrogen and stored at –80°C until further use.Each treatment consisted of three biological replicates.
2.3.Small RNA library construction and deep sequencing
Nematode RNAs were collected separately, and total RNA was isolated using TRIzol Reagent (Ambion, Foster City, CA, USA).Small RNAs (18–30 nt) were isolated by the denaturing PAGE method of Lagos-Quintanaet al.(2001).Agilent 2100 Bio-Analyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) assess the RNA quality.Small RNA sequencing libraries were then constructed using the TruSeq Small RNA Library Preparation Kit (Illumina, San Diego, CA, USA), and Library sequencing was performed by Illumina.
2.4.Preprocessing of microRNAs sequencing data
A general workflow of bioinformatics analyses is shown in Appendix A, including the following steps: (1) Discarding low-quality sequences, reads with a quality score lower than 20 were removed.(2) Discarding reads without 3′ adapter, with 5′ adapter contaminants, without the insert tag.(3) Collecting short RNAs ranging from 15 to 30 nt.Too short (<15 nt) and too long (>30 nt) reads were removed from the analyses.(4) Removing sequences with polyA tails.
2.5.Analysis of M.graminicola miRNAs
We grouped the identical clean reads into unique sequence tags.miRdeep2 Software was used to infer the precursors and the mature sequences of miRNA genes.Only the candidate precursors with hairpinlike structures were kept.The miRNA that targets the sequences ofMgPDIandMgPDI2were predicted using RNAhybrid (Krueger and Rehmsmeier 2006) with default parameters.
2.6.qPCR of mgr-mir-9
miRNAs were extracted from J2 and J3/J4s nematodes using the High Pure miRNA Isolation Kit (Roche, Mannheim, Germany).J3/J4s ofM.graminicolawere collected by dissecting the galls 7 days post-infection (dpi).The quality and concentrations of total RNAs were estimated by electrophoresis and NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, MA, USA).Complementary DNAs (cDNAs) were synthesized using the NCode miRNA First-Strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA).The universal qPCR primer was provided by this kit.The miRNA-specific forward primers (Table 1) were designed based on mgrmir-9 sequences by changing the U in the selected miRNA sequence to T, and the Tm(melting temperature) was adjusted to range from 50 to 55°C.
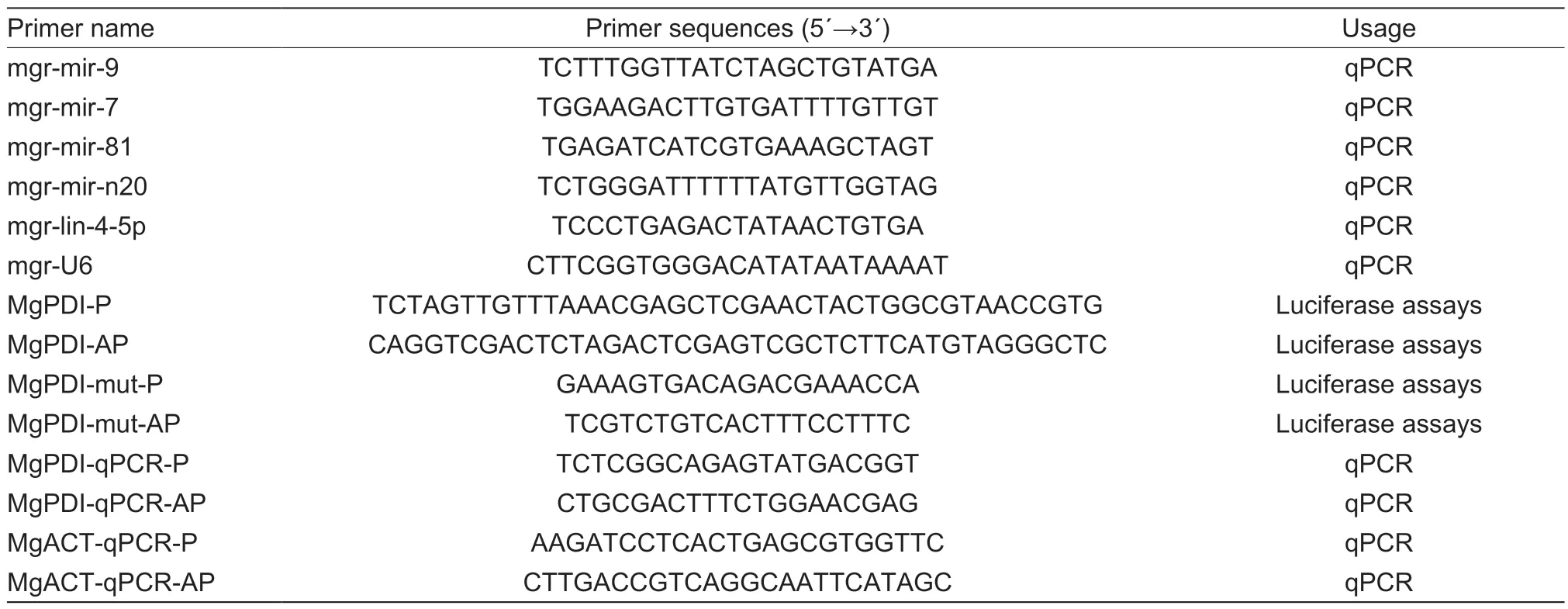
Table 1 List of primers used in this study
The RT-qPCR analyses were conducted using the CFX Connect Real-Time System (Bio-Rad, Hercules, CA, USA) and THUNDERBIRD qPCR Mix (Toyobo, Osaka, Japan).Each RT-qPCR assay was performed at least three biological replicates under the following conditions: 95°C for 60 s and 40 cycles of 95°C for 15 s and 60°C for 30 s.Small nuclear RNA (snRNA) U6 gene ofM.graminicolawas used as the internal control.The RT-qPCR data were analyzed using the 2–ΔΔCtmethod (Livak and Schmittgen 2001).
2.7.Luciferase assays
Target sequences of theMgPDIgene were cloned into the pMIR-REPORT miRNA Expression Reporter Vector (Invitrogen, USA).Mutations at the mgr-mir-9 seed sequence binding region were introduced at the miRNA target sites in theMgPDIgene (GenBank accession number MH392200, primers are indicated in Table 1).The mutant construct was used as the negative control.
Human Embryonic Kidney 293 (HEK 293) cells were bought from Thermo Fisher and has been authenticated by short tandem repeat (STR) profiling, and the cells were free from the mycoplasma contamination.HEK 293 cells were maintained at 37°C in 5% CO2in Dulbecco’s modified Eagle’s medium (D-MEM) (Gibco BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL).Constructs were transfected into HEK 293 cells using Lipofectamine 3000 (Invitrogen).Mimics of mgr-mir-9 were also transfected into the HEK293 cells.The miRCURY LNA miRNA mimic negative control (Exiqon 479903-001, Woburn, MA, USA) was used as a negative control.The Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to measure the interaction between mgr-mir-9 andMgPDI.At 48 h post-transfection, the luciferase activity was measured according to the recommended protocol using a Thermo Scientific Varioskan Flash Microplate Reader (Thermo Fisher Scientific).
2.8.miRNA agomir treatment and infection assay
mgr-mir-9 mimics were synthesized by Sangon Biotech (Shanghai, China).Mimics soaking was performed using the modified method of Rossoet al.(2005) and Huanget al.(2006).A total of 25 000 freshly hatched J2s ofM.graminicolawere soaked in the miRNA solution (0.1 mg mL?1mimics or mimic negative control, 3 mmol L–1spermidine, 50 mmol L–1octopamine, and 0.05% gelatin, adjusted with 0.25× M9 buffer) for 24 h at room temperature in the dark on a rotator.Then, J2s were washed with diethylpyrocarbonate (DEPC) water three times.For detecting the amount of mgr-mir-9 by RTqPCR, miRNAs were extracted from 5 000 treated J2s.To detect the expression level ofMgPDI, total RNA of treated J2s was isolated using the Trizol method (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.The primers are indicated in Table 1.
For the infection assay, Pluronic F-127 (PF-127) (Sigma-Aldrich, St.Louis, MO, USA) gel was used as previously reported (Wanget al.2009; Duttaet al.2011).A total of 80 treated J2s were inoculated on each rice seedling according to Tian Z Let al.(2017).Roots were stained with acid fuchsin (Bybdet al.1983) and the number of eggs was counted after dissecting the stained galls under a microscope at 15 dpi.To determine the reproductive potential ofM.graminicola, a nematode multiplication factor (MF=Number of egg masses×Number of eggs per egg mass/Nematode inoculum level) was also calculated.
2.9.Statistical analysis
All statistical analyses were performed in SPSS Statistics 20.0 Software (IBM).Data had a normal distribution and are presented as means±standard deviation (SD).All data were analyzed by ANOVA (one-way) with a Tukey’s test, with a significance threshold ofP<0.05.
3.Results
3.1.Overview of the small RNA sequencing results
Deep sequencing was used to sequence the three miRNA libraries of J2 juveniles ofM.graminicola.After quality control, 23 528 125, 15 270 859, and 13 082 361 raw reads were obtained from J2a, J2b, and J2c, individually (Appendix B).Raw data are available at NCBI-GEO with accession number: PRJNA824133.After removing contaminants and masking adaptor sequences, we obtained 22 688 911 (96.43%), 14 637 819 (95.85%), and 12 440 375 (95.09%) clean reads (Appendix B).The small RNA length distribution shows abundant sequences in the size range 20 to 29 nt with a peak centered at 23 nt (Fig.1).

Fig.1 Size distribution of the clean reads of Meloidogyne graminicola small RNAs across three libraries.The length distributions of the small RNAs from the three libraries were mainly distributed between 20 and 29 nt and had a peak length of 23 nt.
3.2.ldentification and validation of miRNA of M.gra- minicola
The clean reads were blasted with the miRBase database Release 22.1 (Kozomaraet al.2019) to identify the miRNA genes ofM.graminicola.We also analyzed the hairpin structure, an important structure for folded pre-miRNAs, and removed repeated sequences.A total of 301 conserved miRNAs from the J2a library, 277 conserved miRNAs from the J2b library, and 272 conserved miRNAs from J2c library were identified (Table 2).The conserved miRNAs from at least two libraries were consideredM.graminicolamiRNAs.Consequently, 233 conserved miRNAs belonging to 38 known miRNA families were identified (Table 2).The conserved miRNAs were from 15 to 26 nt in length.We noted a significant biased position of W (56% U or 27% A) at the first position of the miRNAs (Fig.2).Across the three libraries, we identified 21 novel miRNAs (Table 2), which do not have homologs in other nematode species.These 21 novel miRNAs may beM.graminicola-specific.The sequences ofM.graminicolamiRNAs are shown in Tables 3 and 4.
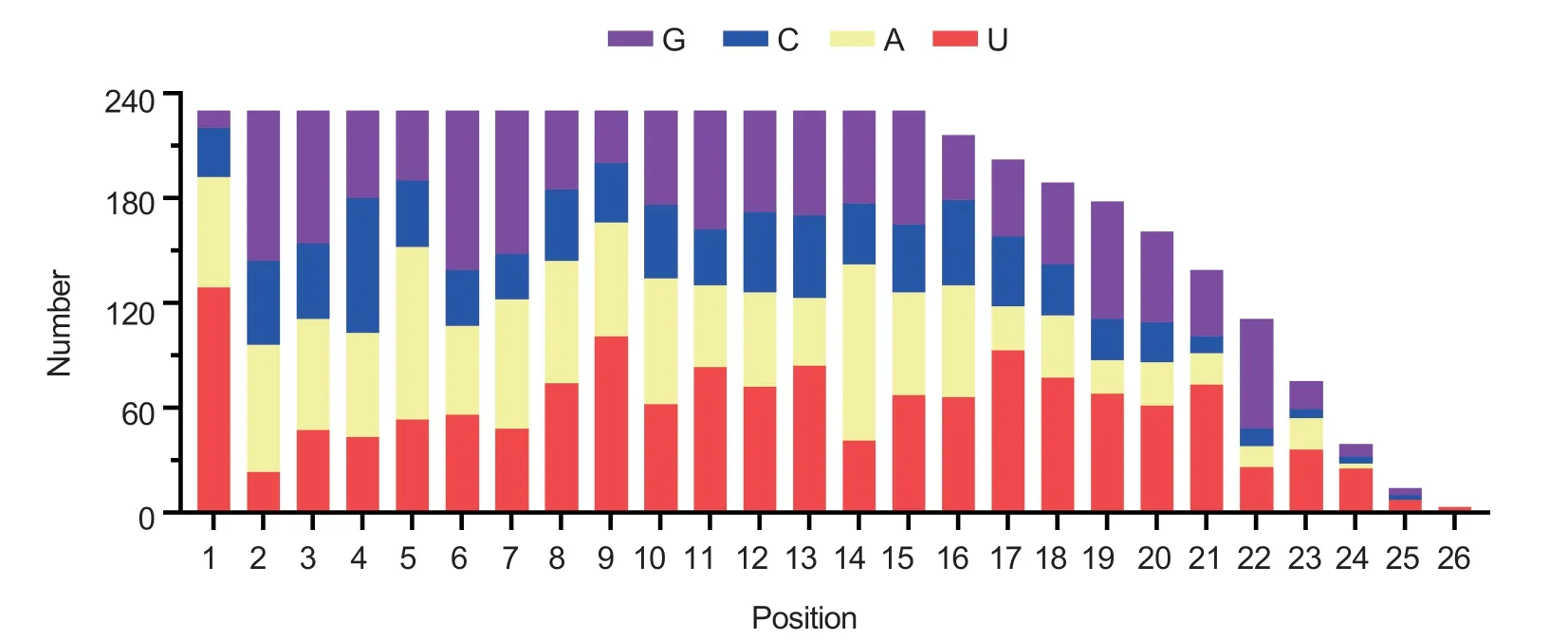
Fig.2 Distribution of bases at each position of Meloidogyne graminicola conserved miRNAs.The first base of the miRNAs tends to be W (U or A).
After counting the reads ofM.graminicolamiRNA, the most abundant 11 miRNAs families were identified in this study, which has more than 500 miRNA sequence reads in all libraries.They are mgr-lin-4, mgr-mir-1, mgrmir-100, mgr-mir-86, mgr-mir-279, mgr-mir-87, mgrmir-71, mgr-mir-9, mgr-mir-50, mgr-mir-72, and mgrmir-34 (Table 3).The most abundant are mgr-lin-4-5p and mgr-mir-100, representing 68.4% of the total miRNA reads in J2a libraries, 69.4% of the total miRNA reads inJ2b libraries, and 68.4% of the total miRNA reads in J2c libraries.The five most abundant novelM.graminicolamiRNAs, mgr-mir-n16, by mgr-mir-n20, mgr-mir-n03, mgrmir-n17, and mgr-mir-n01, with more than 10 000 reads across all libraries were also identified.
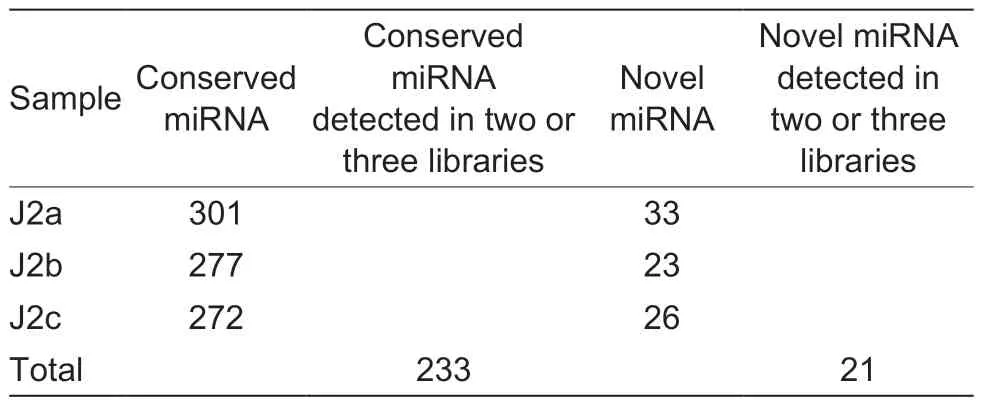
Table 2 The number of Meloidogyne graminicola miRNAs

Table 3 The miRNA families of Meloidogyne graminicola
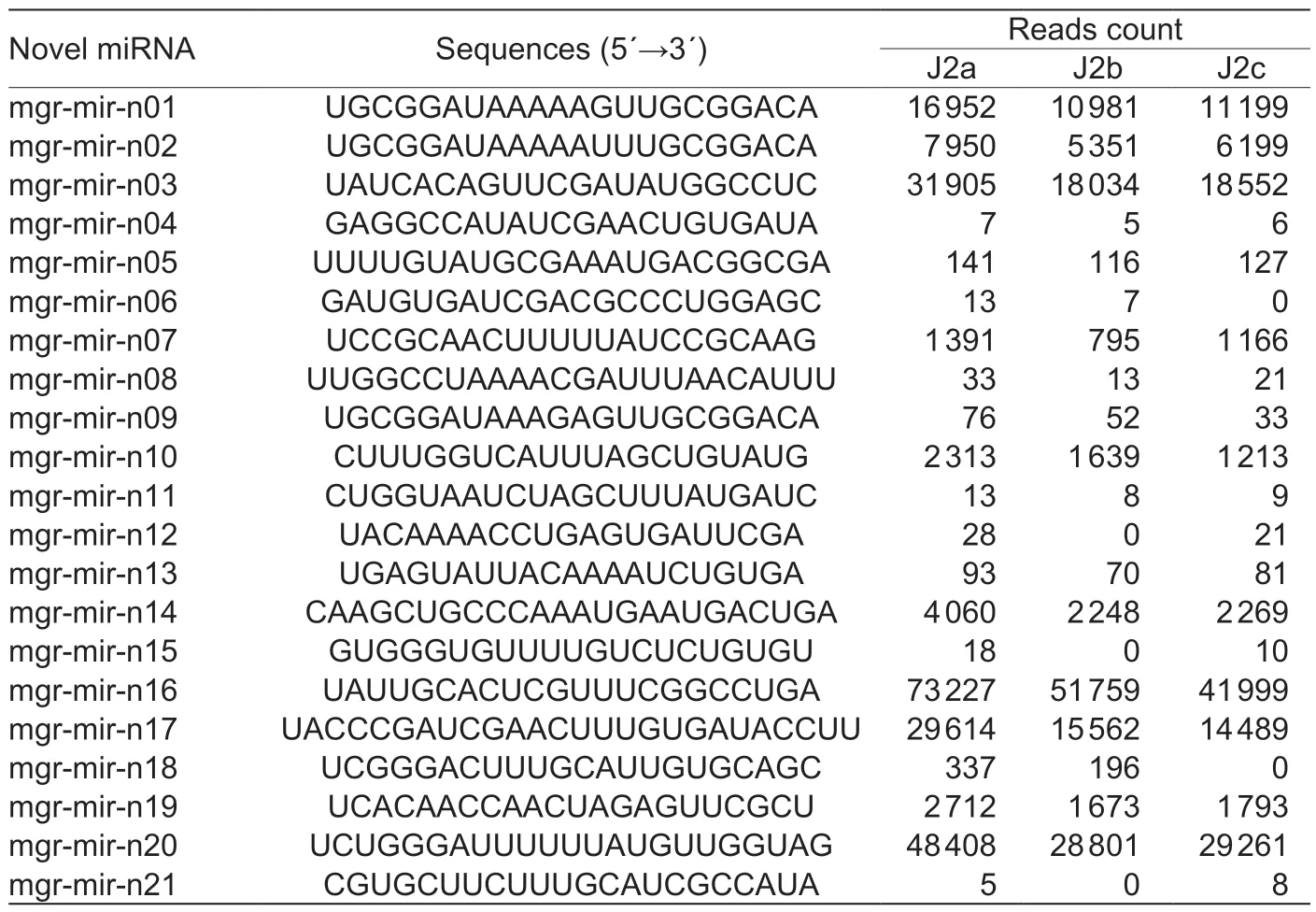
Table 4 The novel miRNA of Meloidogyne graminicola
Five miRNAs, including four conserved miRNAs (mgrlin-4-5p, mgr-mir-7, mgr-mir-9, and mgr-mir-81) and one novel miRNA (mgr-mir-n20), were randomly selected for the validation using RT-qPCR.All selected miRNAs could be detected using RT-qPCR with specific primers.Five selected miRNA expression profiles obtained by deep sequencing correlated well with RT-qPCR data (Fig.3).
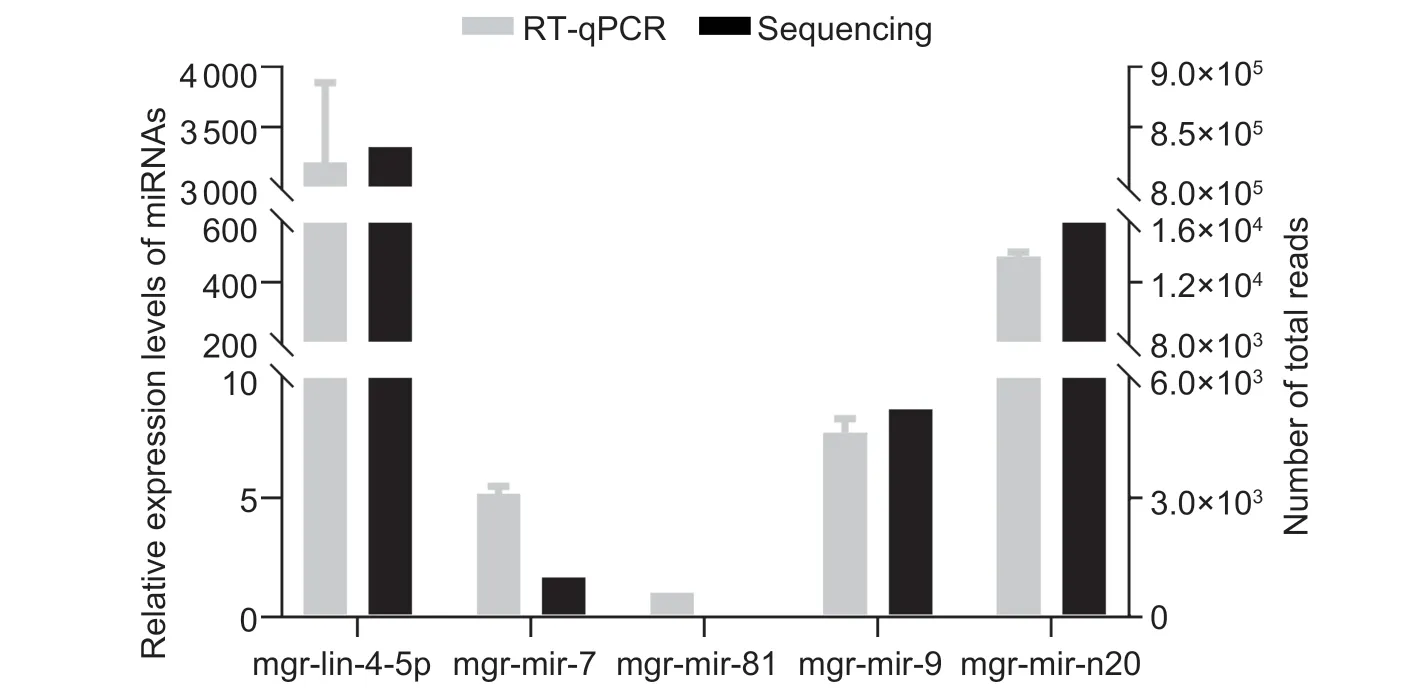
Fig.3 The expression levels of selected Meloidogyne graminicola miRNAs detected by RT-qPCR and by high-throughput sequencing.The expression levels of four miRNAs were in general agreement with the gene expression trends obtained from deep sequencing data.
3.3.Validation of MgPDI mRNA and mgr-mir-9 interaction
DuringM.graminicolainfection, two effector genes,MgPDIandMgPDI2, were reported to be upregulated during the early parasitic stage (Tianet al.2019, 2020).We hypothesized thatM.graminicolamiRNAs enriched at J2 stages could regulate the expressions of secreted effectors includingMgPDIandMgPDI2.To this end, we used a miRNA targets prediction algorithm RNAhybrid to identifyMgPDIandMgPDI2regulators within the miRNA libraries reported in this study.The results showed that mgr-mir-9 could target theMgPDImRNA (Fig.4-A).However, no miRNAs were predicted to target theMgPDI2mRNA.Further, we performedin vitrodual-luciferase reporter assay using anMgPDIconstruct expressed in HEK293 cells to validate the interaction between mgrmir-9 andMgPDImRNA.Results indicated that mgr-mir-9 treatment reducedMgPDIexpression (Fig.4-B).Further, we detected the expression level ofmgr-mir-9at the J2 stage (0 dpi) and J3/J4 stage (7 dpi).The results showed that the expression level ofmgr-mir-9was greatly reduced compared to the level right before infection (Fig.4-C).The expression trend ofmgr-mir-9was contrary to the expression level ofMgPDIat the J2 stage (0 dpi) and J3/J4 stage (7 dpi) (Tianet al.2019).Together, these results suggest that mgrmir-9 could down-regulate the expression level of its targetMgPDI, and at 7 dpiMgPDIwas upregulatedviathe reduced expression ofmgr-mir-9.

Fig.4 The interaction between mgr-mir-9 and MgPDI mRNA.A, pairing schemes of mgr-mir-9 and MgPDI mRNA interactions are illustrated.The matching position of MgPDI mRNA starts at 879 in the open reading frame.B, MgPDI is direct target of mgr-mir-9.pMIR-REPOR-MgPDI luciferase constructs containing a wild type (WT) or mutant type (MT) target sequence were transfected into HEK293 cells.Relative fluorescence ratios in HEK293 cells co-transfected with the indicated pMIR-REPORT-MgPDI luciferase vector plus either a negative control miRNA (NC), or mgr-mir-9 mimics.Results showed the mean values of four independent experiments±SD (n=4).C, the expression level of mgr-mir-9 at the J2 and J3/J4 stages.Vertical bars represent the mean±SD (n=3).All data were analyzed by ANOVA (one-way) with a Tukey’s test (*, P<0.05; **, P<0.01).
3.4.mgr-mir-9 involved in M.graminicola infection in rice
To determine the effects of mgrmir-9 on the target geneMgPDIin vivo, we soaked fresh J2s in mgr-mir-9 mimics or mimics negative control for 24 h.The accumulation ofmgr-mir-9was detected using RT-qPCR.The results showed that the level of mgr-mir-9 after soaking was significantly higher compared with the control (Fig.5-A).As expected, the expression level of its target,MgPDI, was downregulated (Fig.5-B).There are no differences about the number of root knots and egg masses between the treated and control groups according to the infection studies.However, the treatment of J2s with mgrmir-9 mimics significantly reduced the reproductive ability of nematodes (MF=7.08)compared to the mimics negative treatment (MF=10.82) (Fig.5-C).Above all, these suggested that mgr-mir-9 may play an important role.

Fig.5 mgr-mir-9 involved in nematode infection.A, the relative accumulation level of mgr-mir-9 at 24 h post different treatments.B, the relative expression level of MgPDI at 24 h post different treatments.C, the multiplication factor values at 15 d post infection (dpi).NC, treated with a negative control miRNA; mimics, treated with mgr-mir-9 mimics.Vertical bars represent the mean±SD (n=16).All data were analyzed by ANOVA (one-way) with a Tukey’s test (*, P<0.05; **, P<0.01).
4.Discussion
The RKN,M.graminicola, is an important parasite that attacks rice and causes substantial destruction in most rice-growing areas (Pokharelet al.2010; Mantelinet al.2017).Like other plant hosts, rice utilized a multilayered innate immune system against nematode infection (Jones and Dangl 2006).However, during the long-term coevolution, plant-parasitic nematodes (PPNs) have adapted a sophisticated strategy to avoid triggering a host intense defense response.Directly delivering effectors through stylet into the apoplast and cytoplasm of host cells is a unique strategy of PPNs to suppress host defense responses (Goverse and Smant 2014).Most research focused on the function of nematode effectors.Here we reported RKN miRNAs, as important gene regulators, are involved in the important effector expression regulation during early stages of the infection.
During the interactions between plant-parasitic nematode and hosts, the plant miRNAs play an important role.For example, in rice, differentially expressed miRNAs responsive to early infection withM.graminicolahave been identified (Verstraetenet al.2021).However, to our knowledge, studies on the roles ofM.graminicolamiRNA have not been reported yet.This study identified conserved and novelM.graminicolamiRNAs by deep sequencing.Nearly 52 million raw reads were acquired.In total, 233 conserved miRNAs and 21 novel miRNAs were identified.TheM.graminicolamiRNAs had a peak length of 23 nt, similar to those ofM.incognita(Wanget al.2015).BothM.graminicolaandM.incognitamiRNAs have a significant bias of U or A at the first position (Wanget al.2015).Among the identifiedM.graminicolamiRNAs, the reads counts of mgr-lin-4-5p and mgr-mir-1 are much more than other miRNAs.miR-1,a conserved muscle-specific miRNA, was reported to regulate both pre- and post-synaptic function atC.elegansneuromuscular junctions (Simonet al.2008), and lin-4 was reported to control the timing ofC.eleganslarval development (Wightmanet al.1993).mgr-lin-4-5p and mgr-mir-1 may have similar functions inM.graminicola.Besides the known miRNA families, we also identified 21 novel miRNAs.The functions of theseM.graminicolaspecific miRNAs need further study.
Emerging evidence suggests that miRNAs play a major role in virtually all biological and metabolic processes, including growth, reproduction, organ differentiation, disease pathogenesis, and even responses to stressors (Zhang and Wang 2015).Mani (2021) confirmed that specific miRNAs targeting different mRNAs was expressed at different development stages inM.incognita(Maniet al.2021).However, those differently expressed miRNA’s physiological and biological functions during PPNs and host interactions need further study.In this study, a class of differently expressedM.graminicolamiRNAs were identified at the J2 stage.Moreover, we aimed to elucidate the effector mRNA targets ofM.graminicolamiRNAs and foundMgPDImRNA is one of the targets.The PDI is a member of the thioredoxin superfamily, which can catalyze the formation, reduction, and isomerization of disulfide bonds of proteins (Sevier and Kaiser 2002).The PDIs of pathogens act as an important virulence factor during host infection (Stolfet al.2011).
Our previous studies identified twoPDIgenes inM.graminicola,MgPDIandMgPDI2(Tianet al.2019, 2020).MgPDI was involved in protection against oxidative damage, while MgPDI2 could induce strong necrotic responses inNicotianabenthamiana(Tianet al.2019, 2020).It is speculated that the timing of the effectors’ expression and secretion must be precisely regulated.miRNA could play a vital role in the effector regulation.Predicted by RNAhybrid and validated byinvitroluciferase assays, mgr-mir-9 andMgPDImRNA interaction was first reported.And a negative correlation between expression profiles of mgr-mir-9 andMgPDIwas also confirmedinvivo.Moreover, exogenous mgrmir-9 treatment ofM.graminicolaJ2s could significantly suppress the expression ofMgPDI.mgr-mir-9 mimics treatment significantly reduced the reproductive ability of nematodes (MF=7.08) compared to that of the treatment of J2s with negative control (MF=10.82).Taken together, mgr-mir-9 is involved in theM.graminicolainfection process by removing the inhibition on its effector target,MgPDI, once inside the host.
The mir-9 family was also identified in other PPNs such asBursaphelenchusxylophilus(Dinget al.2015) andM.incognita(Wanget al.2015), while its target was still unknown.InM.graminicola, the mir-9 family contains three genes, which transcribe mgr-mir-9a (UCUUUGGUUAUCUAGCUGUAUGA), mgr-mir-9b (UCUUUGGUUAUCUAGCUGUAUG) and mgr-mir-9c (UCUUUGGUUAUCUAGCUGUAU).The mgr-mir-9a is the most abundant form, while the mgr-mir-9c is the least.miRNA members among mir-9 have high homology in sequence, which makes it difficult to be discriminated.
5.Conclusion
We identified 233 known miRNAs and 21 novel miRNAs ofM.graminicolathrough deep sequencing, which provides an overview ofM.graminicolamiRNAs at early stages for the first time.More importantly, we found that mgr-mir-9 was involved in the infection ofM.graminicolaby down-regulating its target,MgPDI.Present work is the first to elucidate the role of miRNAs inM.graminicolainfecting rice and provides new insights that nematode non-coding RNAs can be used as a potential resource for the development of new crop protection strategies.We anticipate detailed functional studies on other plant–nematode miRNAs will help to reveal the interactions between plants and nematodes and may aid in devising effective plant protection strategies against parasitic nematodes.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (32001877).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2022.08.127
 Journal of Integrative Agriculture2023年5期
Journal of Integrative Agriculture2023年5期
- Journal of Integrative Agriculture的其它文章
- Herbicidal activity and biochemical characteristics of the botanical drupacine against Amaranthus retroflexus L.
- Developing a duplex ARMS-qPCR method to differentiate genotype l and ll African swine fever viruses based on their B646L genes
- The effects of maltodextrin/starch in soy protein isolate–wheat gluten on the thermal stability of high-moisture extrudates
- Elucidation of the structure, antioxidant, and interfacial properties of flaxseed proteins tailored by microwave treatment
- Effects of planting patterns plastic film mulching on soil temperature, moisture, functional bacteria and yield of winter wheat in the Loess Plateau of China
- lnversion tillage with straw incorporation affects the patterns of soil microbial co-occurrence and multi-nutrient cycling in a Hapli-Udic Cambisol
