Establishing Gene Delivery Systems Based on Small-Sized Chitosan Nanoparticles
WANG Liping , ZHANG Wenhua , ZHOU Quan , LIU Shihai,XU Wenhua SUN Teng, and LIANG Ye
1) Key Laboratory, Department of Urology and Andrology, Affiliated Hospital of Qingdao University,Qingdao 266003, China
2) Department of Inspection, Qingdao University, Qingdao 266003, China
3) Central Laboratory, Affiliated Hospital of Qingdao University, Qingdao 266003, China
4) Laboratory Animal Center, Affiliated Hospital of Qingdao University, Qingdao 266003, China
5) Department of Outpatient, Affiliated Hospital of Qingdao University, Qingdao 266003, China
(Received September 6, 2017; revised December 13, 2017; accepted March 2, 2018)
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2018
Abstract Chitosan is a natural cationic polysaccharide, which is often used for preparing biomedical materials because of its high biocompatibility. In this study, chitosan with a molecular weight of 160 kDa was chosen to prepare chitosan nanoparticles (CSNPs)as gene vectors by ionic cross-linking with tripolyphosphate (TPP). CSNPs were characterized in terms of particle size, zeta potential,and polydispersity index (PDI) using a Zetasizer, and morphology was evaluated by transmission electron microscopy (TEM). Furthermore, the cytotoxicity and biocompatibility of CSNPs were correspondingly examined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and histological examination. Agarose gel electrophoresis and UV spectrophotometric methods were performed to measure the loading capacity. The cell transfection efficiency of CSNPs loaded with plasmids or siRNA was analyzed by fluorescence microscopy or laser scanning confocal microscopy. The results showed that CSNPs were prepared successfully by the ionic gelation method, which had a smaller particle size (100 nm -200 nm), stable dispersibility, low cytotoxicity,good tissue-biocompatibility, and high gene-loading efficiency. These CSNPs could transfer the plasmids or siRNA to cells. However, CSNPs might have a much higher transfection efficiency for siRNAs than for plasmids, which implies that CSNPs might be a safer and more efficient vector for delivering siRNAs rather than plasmids.
Key words chitosan; nanoparticles; gene delivery
1 Introduction
Gene delivery systems have recently attracted much attention in medical, pharmaceutical, and biotechnological fields owing to their applications in disease treatments(Ewert et al., 2005; Storrie and Mooney, 2006). However,the most important problem in gene therapy is to find a suitable vector that is safe and efficient. Recently, studies have focused on polymeric nanoparticles, because they can improve the therapeutic efficacy of drugs by regulating their release and prolonging circulation time (Sung et al., 2012; Jain et al., 2016).
Chitosan (CS) is derived from chitin, an abundant biopolymer that is mostly found in the exoskeletons of crustaceans and insects, as well as in the cell walls of fungi(Singla and Chawla, 2001; Pillai et al., 2009). Because it contains abundant amino and hydroxyl groups, chitosan is capable of forming nanoparticles via both physical and chemical cross-linking (Liu et al., 2005; Makhlof et al.,2011). Chitosan can be cross-linked with negatively charged multivalent ions, such as tripolyphosphate (TPP),by non-covalent interactions to form nanoparticles (Liu et al., 2007; Tsai et al., 2011). Owing to the presence of glucosamine groups on the backbone of chitosan, it possesses a high density of positive charge, which can easily mediate binding with the negative charge of biomolecules such as oligonucleotides (Howard et al., 2006; Puras et al.,2013), therefore, chitosan can be used as a gene carrier material.
Chitosan nanoparticles (CSNPs) are a potential drug delivery system because of their high biocompatibility,low cytotoxicity, and immunogenicity (Hu et al., 2006;Qian et al., 2006; Reverchon and Antonacci, 2006; Dash et al., 2011). Furthermore, many studies indicate that CSNPs have great potential for the targeted and efficient delivery of cancer treatment drugs (Chowdhuri et al.,2016; Li et al., 2016; Shen et al., 2016). However, there are few reports on which kind of gene products could be suitable for delivery by CSNPs of such small size.
In this following study, chitosan with a molecular weight of 160 kDa was chosen to formulate nanoparticles by cross-linking with TPP, using the ionic gelation method. Suitable CSNPs were selected by characterization in terms of particle size, zeta potential, and polydispersity index, and morphology was evaluated by transmission electron microscopy (TEM). Furthermore, the cytotoxicity and biocompatibility of the CSNPs were evaluated,and the cell transfection efficiency of plasmid- or siRNA-loaded CSNPs was assessed to determine the gene product most efficiently delivered by CSNPs.
2 Materials and Methods
2.1 Materials
Chitosan (MW 160 kDa, deacetylation degree of more than 90%), sodium tripolyphosphate (TPP), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),and Lip3000 were purchased from Sigma-Aldrich (St.Louis, MO, USA). Kunming mice were purchased from Qingdao Laboratory Animal Center (Qingdao, China).A549 and L929 cell lines were supplied by the cell bank of the Chinese Academy of Sciences, Beijing, China.Materials for cell culture, including RPMI1640 culture medium, DMEM culture medium, fetal bovine serum(FBS), trypsin, penicillin, and streptomycin were purchased from GIBCO (Grand Island, NY, USA). Plasmids(pIRES2-EGFP) were prepared onsite in the laboratory.siRNA was purchased from Shanghai Sangon Biotechnology Co. (Shanghai, China). HiPerFect transfection reagent was purchased from QIAGEN (Hilden, Germany).All other reagents used were of reagent grade.
2.2 Methods
2.2.1 Preparation of CSNPs
CSNPs were prepared according to the described method (Calvo et al., 1997). CS solutions were prepared at concentrations of 1 mg mL-1and 2.5 mg mL-1by dissolving CS in a 0.5% acetate buffer solution. The TPP was dissolved in double distilled water to concentrations of 1 mg mL-1and 2.5 mg mL-1. All solutions were passed through a 0.22-μm filter. CSNPs were prepared by adding TPP drop-wise to the CS solution under stirring, with a CS:TPP ratio of 10:1 or 5:1 (w/w). The mixtures were left stirring for 30 min at room temperature, and then subjected to a low-frequency ultrasonic treatment for 2 min.
2.2.2 Size and zeta potential measurement
The mean particle size, polydispersity index (PDI), and zeta potential of the CSNPs were analyzed by a Zetasizer(Nano ZS; Malvern Instruments, Malvern, UK) to screen the optimal size of nanoparticles for subsequent experiments.
2.2.3 Transmission electron microscopy observation
The morphological characterization of CSNPs was determined by transmission electron microscopy (TEM)(JEM-1200EX; JEOL, Tokyo, Japan). Briefly, the freshly generated nanoparticles were dried on a copper disk, and then examined by TEM.
2.2.4 Cytotoxicity detection
The cytotoxicity of CSNPs was tested with a monolayer of L929 cells according to ISO standards (GB/T 16886.5-2003/ISO 10993-53199). CSNPs were prepared and dialyzed overnight using a semipermeable membrane(intercept MW, 100 kD), and then filtered by a 0.22-μm filter. Then, 5 × 103cells were seeded on each well of 96-well culture plates in a DMEM medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin(100 IU mL-1). After being cultured for 24 h at 37℃ in a 5% carbon dioxide atmosphere, cells were treated with increasing amounts of CSNPs (5 μL, 20 μL, and 50 μL),and compared to non-treated control cells and the corresponding CS solution. Cell viability was assessed after 24 h, 48 h, and 72 h treatments by MTT assay. All experiments were performed in triplicate. The final results of the OD490 value were analyzed by the relative growth rate (RGR) with the following formula: RGR = ODt/ODc× 100%, where ODt is the value of treatment groups, and ODc is the value of the control. Cellular responses were scored as 0, 1, 2, or 3, corresponding to non-cytotoxic(RGR ≥ 100%), slightly cytotoxic (RGR = 75-99%), moderately cytotoxic (RGR = 50-74%) and severely cytotoxic (RGR = 25-49%), respectively.
2.2.5 Biocompatibility evaluation
Sterile CSNPs were obtained by the same method as described in 2.2.4. Mice were treated with CSNPs by intramuscular injection on the left leg, and compared to the phosphate-buffered saline (PBS) control. After injecting for 1 w, 2 w, and 3 w, the mice were euthanized and the leg muscles were collected. The immune responses were examined by a microscope after hematoxylin & eosin(H&E) staining.
2.2.6 Encapsulation of plasmids or siRNA in CSNPs
Gene loading of CSNPs was performed through the ionic gelation of the chitosan with a mixture of plasmid or siRNA and TPP solution. Briefly, plasmid (pIRES2-EGFP, 700 ng μL-1) or siRNA-Cy3 (20 μmol L-1) to target Bcl-2 were added to the TPP solution (200 μL, 1 mg mL-1)before being added drop-wise to the chitosan solution (2 mL, 1 mg mL-1) under stirring. Then, the mixtures were stirred for 30 min and ultrasonicated for 2 min. The mean particle size, PDI, and zeta potential were analyzed by the Zetasizer.
2.2.7 Loading capacity determination
The loading capacities of the plasmid nanoparticles(CSpNPs) and siRNA nanoparticles (CSsNPs) were determined by agarose gel electrophoresis and UV spectrophotometry. To investigate the gene-binding capacity of the nanoparticles, increasing amounts of plasmids (7-140 μg) or siRNA (0.375-3 OD) were loaded into the CSNPs.After that, the samples were centrifuged at 13000 rmin-1for 30 min, and the supernatant was analyzed by agarose gel electrophoresis to determine the free plasmid or siRNA concentration compared to the initial concentrations. The loading efficiency of 140 μg plasmid and 3 OD siRNA to CSNPs was determined by UV spectrophotometry, and calculated using the following formula:
Loading efficiency = (1 - ODs/ODt) × 100%,
where ODs is the absorbance measurement at 260 nm of plasmid in the supernatant, and ODt is the absorbance measurement at 260 nm of initial plasmid.Jolla, CA, USA). Results were considered significant at P< 0.05 and highly significant at P < 0.01.
2.2.8 Transfecting CSpNPs and CSsNPs to cells
To investigate the cellular uptake abilities of CSpNPs or CSsNPs, different amounts of GFP-plasmid (14, 35, 70,and 140 μg) and siRNA (0.375, 0.75, 1.5, and 3 OD) were loaded onto the CSNPs. A549 cells were seeded on the glass slides in 24-well culture plates with RPMI1640 medium supplemented with 10% fetal bovine serum. After being cultured for 24 h, the culture medium was removed and replaced by medium containing either CSpNPs or CSsNPs, to be compared with non-treated control cells.The Lip3000 and HiPerFect transfecting agents were used as the positive controls. The same amounts of plasmid or siRNA were transfected for each experiment and the corresponding positive control. After culturing with CSpNPs for 48 h or with CSsNPs for 24 h, cells were observed by fluorescence microscope (Olympus, IX71, Tokyo, Japan)or washed by PBS (pH 7.4) three times, and then fixed with formaldehyde solution (4%) for the immunofluorescence assay. After that, cells were analyzed by laser scanning confocal microscopy (SPE; Leica, Heidelberg,Germany).
3 Results
3.1 Characterization of CSNPs
Through analyzing the mean particle size, PDI, and zeta potential of nanoparticles with the Zetasizer, smallsized nanoparticles (100-200 nm) could be prepared by CS (MW 160 kD) and exhibited a narrow range of distribution (PDI < 0.2) (Table 1). The particle size increased along with the CS solution concentration. The ratio of CS to TPP affected the zeta potential, but not the particle size.The nanoparticles formulated with a CS-TPP ratio of 10:1(w/w) showed a higher and more stable zeta potential than the 5:1 (w/w) nanoparticles. Therefore, the CSNPs prepared with a CS (160 kDa, 1 mg mL-1) to TPP ratio of 10:1 (w/w) were used for the subsequent experiments,based on their smaller size, narrow range of distribution,and stable zeta potential. Moreover, TEM showed the CSNPs to be about 100-200 nm in size (Fig.1) and randomly dispersed and distributed, and to have a spherical morphology.
2.2.9 Statistical analysis
Data are shown as mean ± SD of a representative point from triplicate experiments. Statistical analysis of data was performed by Student’s t-test or one-way analysis of variance (ANOVA) using Prism software (GraphPad, La
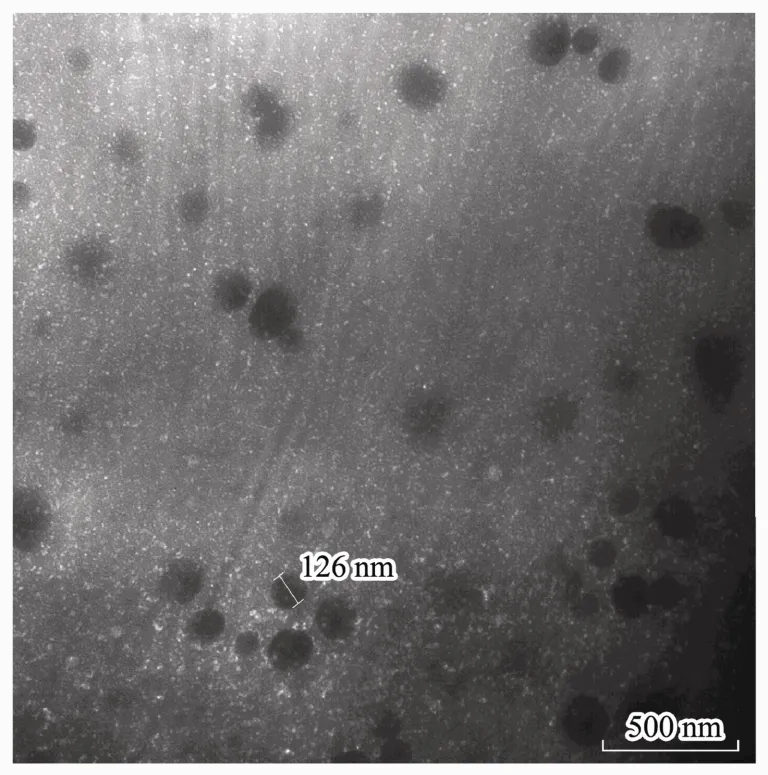
Fig.1 Morphological characteristics of the CSNPs was observed by TEM, and the nanoparticles were almost 100-200 nm in size.
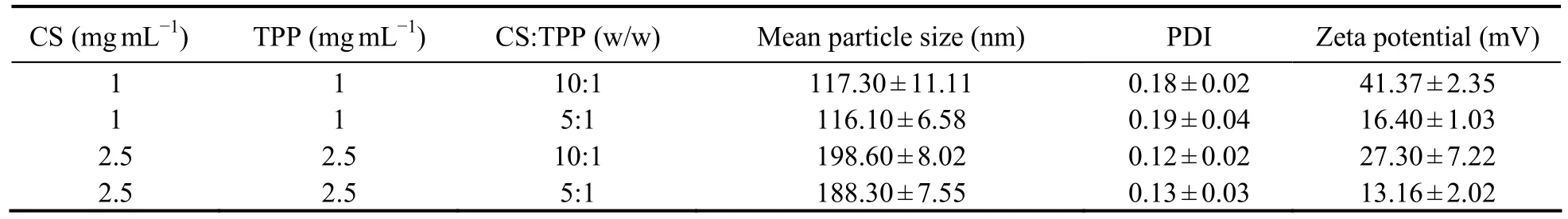
Table 1 Effects of chitosan (CS) solution concentration, TPP concentration, and CS:TPP ratio (w/w) on the mean particle size, polymer dispersity index (PDI), and zeta potential of chitosan nanoparticles (CSNPs)
3.2 Cytotoxicity of CSNPs
CSNPs cytotoxicity at 24 h, 48 h, and 72 h was assessed by MTT assay, and L929 cells cultured with CS solution were used as the control. The results are shown in Fig.2.As the dose of CSNPs increased, there was little influence on the cell viability. All CSNPs groups showed slight cytotoxicity (RGR: 75%-99%), which meets the application requirement for medical materials. However, the CS treatment with the highest dose showed the lowest RGR from 24 h to 72 h, which suggests that the cytotoxicity of CSNPs is lower than that in the CS solution control group.
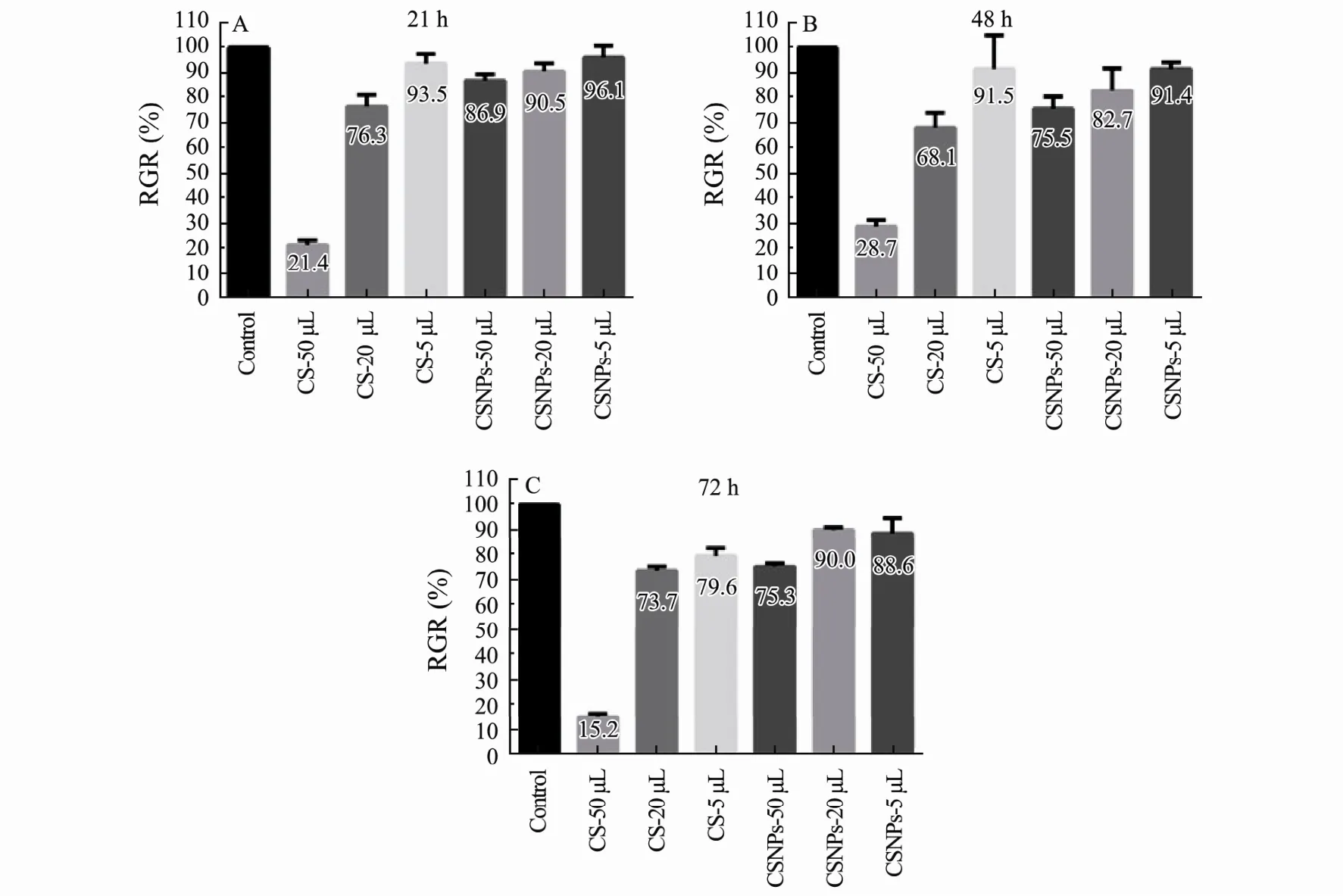
Fig.2 In vitro cytotoxicity was evaluated by MTT assay after culturing cells with CS solution and CSNPs, at 24 h (A), 48 h(B), and 72 h (C). The data are shown as means ± SD of triplicate experiments.
3.3 Biocompatibility of CSNPs
In order to investigate the biocompatibility, mice were treated with CSNPs by intramuscular injection, and the histological findings were assessed with a microscope. As shown in Fig.3, all the groups maintained the normal tissue construction and there were no toxic effects of CSNPs on the muscles of mice compared to that in the PBS control. CSNPs did not cause any tissue damages or obvious inflammation in the muscles of mice after injecting for 1 w, 2 w, and 3 w. These results indicate that the CSNPs show good biocompatibility.
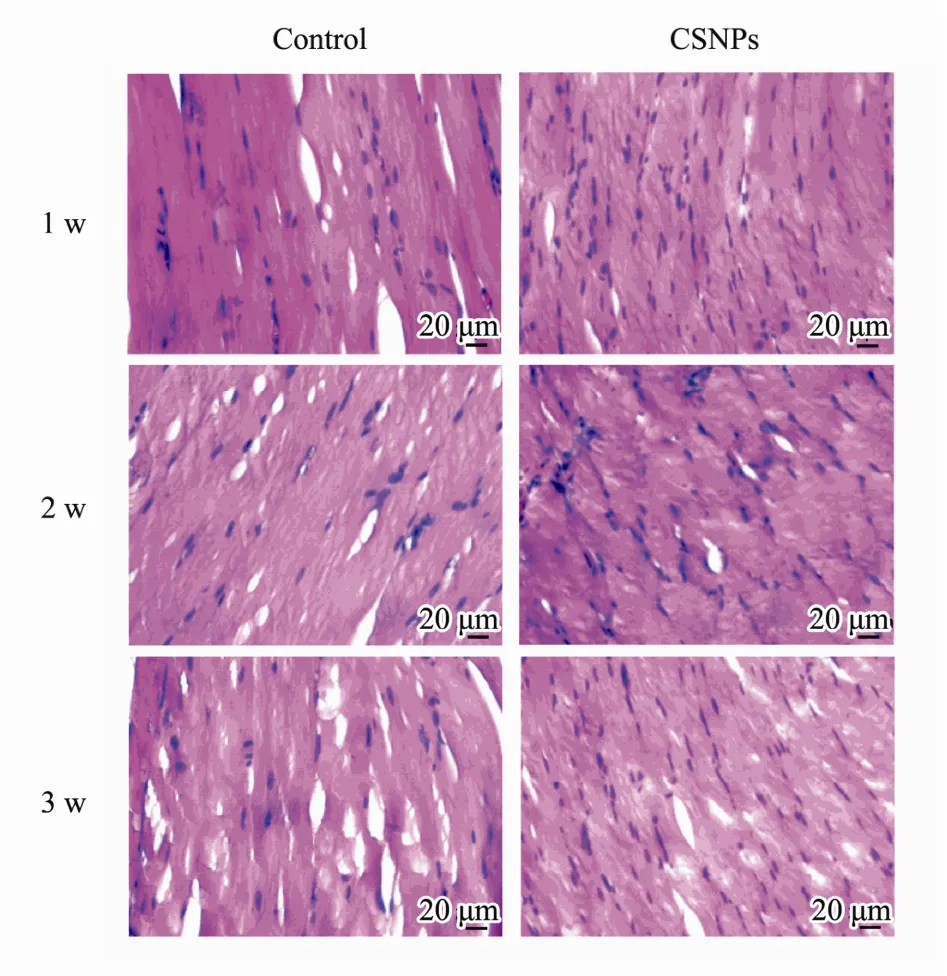
Fig.3 Slides for the histological examination were stained with hematoxylin and eosin (H&E) and assessed with a microscope (400×). Compared to that in the control,CSNPs did not cause obvious inflammation.
3.4 Plasmids- or siRNA-Loaded CSNPs
CSpNPs and CSsNPs were generated by premixing plasmids or siRNA with TPP before adding to the CS solution. Compared to that for the blank CSNP control,the mean particle size, PDI, and zeta potential results are shown in Table 2. The size of the CSpNPs and CSsNPs were slightly larger than the blank CSNPs, and the PDI and zeta potential did not significantly change. Moreover,the size distributions of CSpNPs and CSsNPs were almost the same as the blank nanoparticles, which had a uniform size of almost 100-200 nm (Fig.4).
3.5 Determination of Loading Capacity
To evaluate the loading capacity of CSpNPs and CSsNPs, the amount of unencapsulated plasmids or siRNA was measured by agarose gel electrophoresis and UV spectrophotometry, comparing with the initial plasmid concentration. The results indicate that no free plasmids or siRNA were detected in the agarose gel, while the encapsulated amounts in CSNPs were increased from 7-140 μg for plasmids and 0.375-3 OD for siRNA (Fig.5). UV spectrophotometry results showed that the loading efficiency of CSNPs was more than 90%, even if 140 μg plasmid or 3 OD siRNA were loaded (Table 2). The results all indicate that CSpNPs and CSsNPs have a high capacity for both plasmid and siRNA encapsulation.
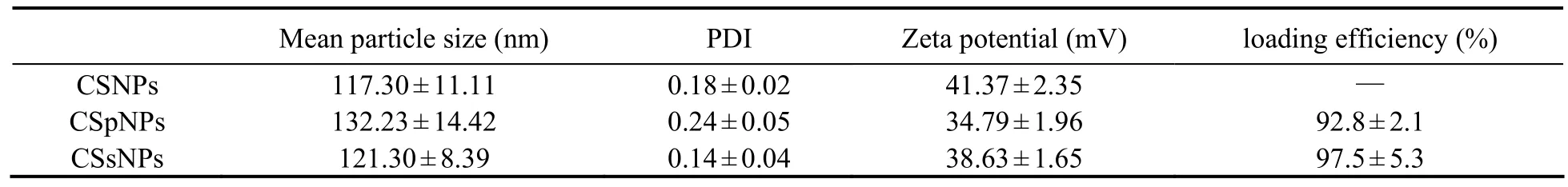
Table 2 The mean particle size, polymer dispersity index (PDI), zeta potential, and loading efficiency of the CSpNPs and CSsNPs compared to the blank CSNPs
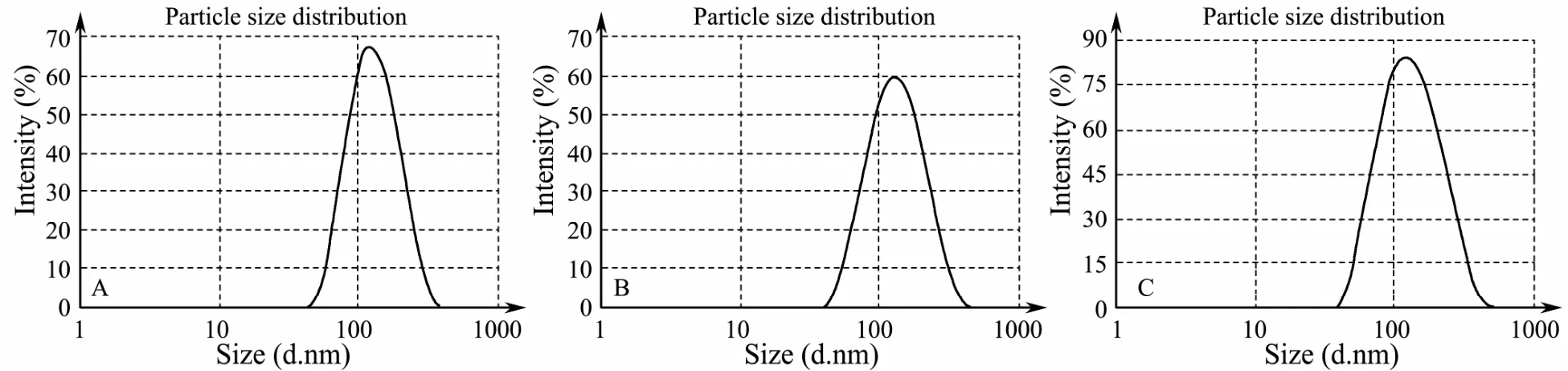
Fig.4 Particle distribution of CSpNPs (B) and CSsNPs (C) compared to that of the blank CSNPs (A).
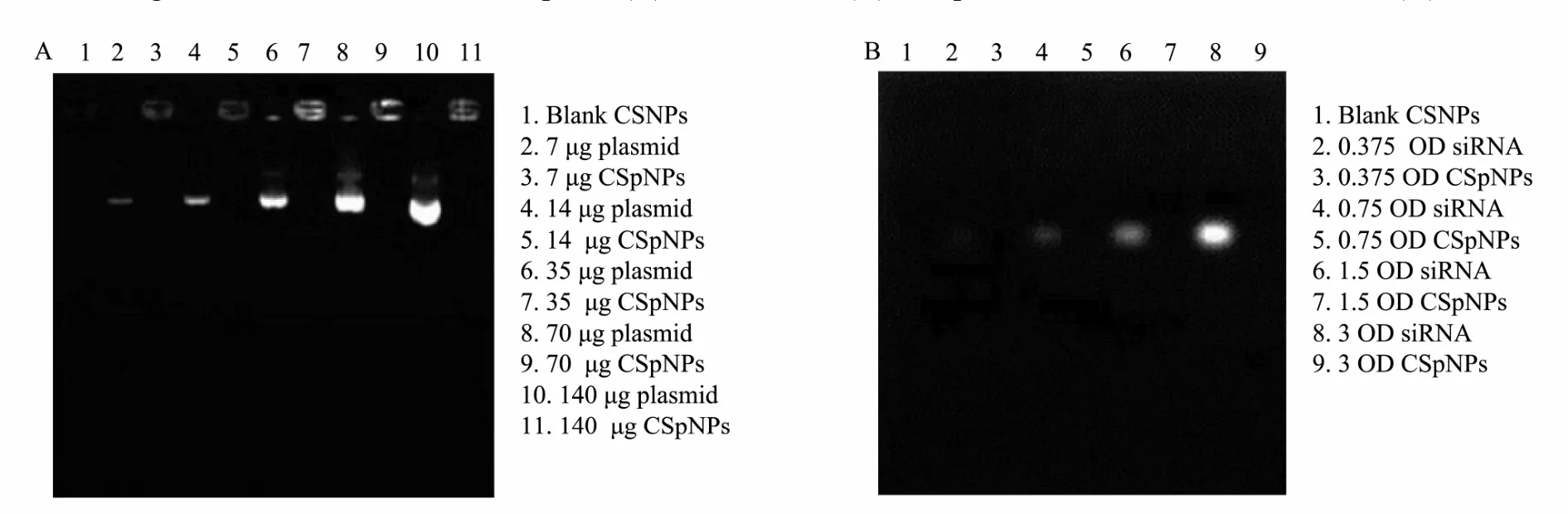
Fig.5 Electrophoresis images of CSpNPs shown on agarose gel. No free plasmids (A) or siRNA (B) were detected whenthe amounts of plasmids or siRNA encapsulated in CSNPs were increased.
3.6 Cell Transfection with CSpNPs
After transfecting for 48 h with GFP-containing plasmid (pIRES2-EGFP)-loaded CSNPs, the GFP expression in A549 cells was investigated. The green fluorescence originating from the plasmids was detected by fluorescence microscopy. As shown in Fig.6, there was no expression of green fluorescence in the non-treated control cells, and some green fluorescence could be observed in cells that were treated with CSpNPs. For the experiment group, the cell fluorescence was enhanced as the plasmid loading amounts increased, which indicated that CSpNPs can transfer plasmids to cells. However, the green fluo-rescence of the experimental group was much weaker than that in the control group of Lip3000, which showed that the transfection rate was much lower than that for the positive control.
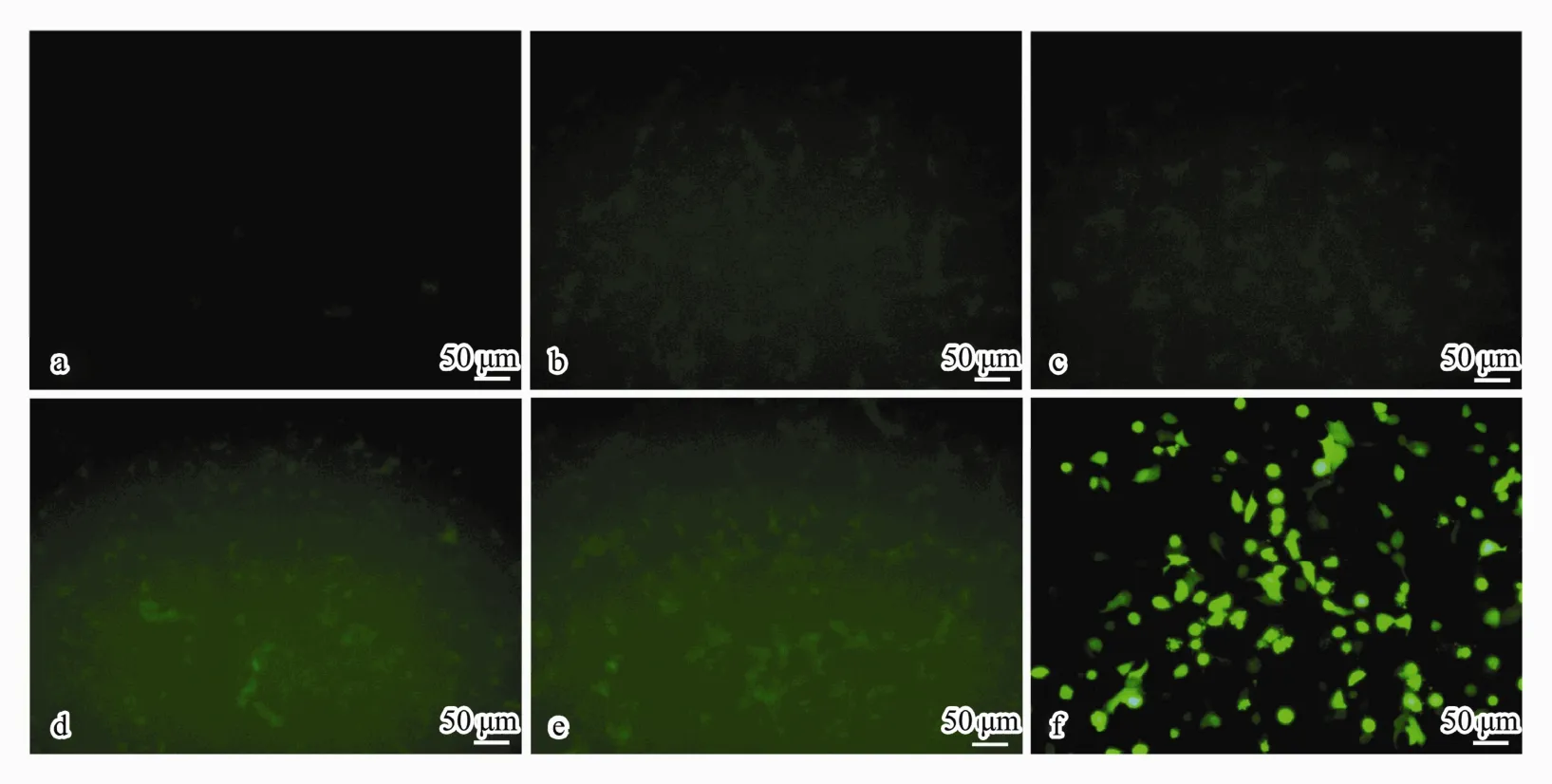
Fig.6 Images of GFP expression in A549 cells observed by fluorescence microscope after treatment with CSpNPs loaded with different amounts (14, 35, 70, and 140 μg) of GFP-plasmid (b-e) compared to that in the normal (a) and positive (f)controls (200×).
3.7 CSsNP-Transfected Cells
Twenty-four hours after treatment with different amounts (0.375, 0.75, 1.5, 3 OD) of Cy3-labeled Bcl-2 siRNA (shows red fluorescence) loaded in CSNPs, fluorescent images of A549 cells were observed with a fluorescence microscope (Fig.7b-e), taking the non-treated cells as the normal control (Fig.7a) and HiPerFect transfection cells as the positive control (Fig.7f). There was no red fluorescence observed in the normal cells. Red fluorescence could be observed in the CSsNPs-treated groups,which was enhanced when the siRNA loading amount was increased. Additionally, the images of CSsNPs loaded with 1.5 OD siRNA in A549 cells were observed by laser scanning confocal microscopy and compared to those for the normal and positive controls (Fig.8). The cytoskeleton was stained with actin (green fluorescence),and the nuclei were stained with DAPI (blue fluorescence). Results showed that the CSsNPs could transfer siRNAs to cells successfully, at a higher transfection rate than that for the positive control (Fig.8), which indicated that CSsNPs could transfect siRNAs with high efficiency.
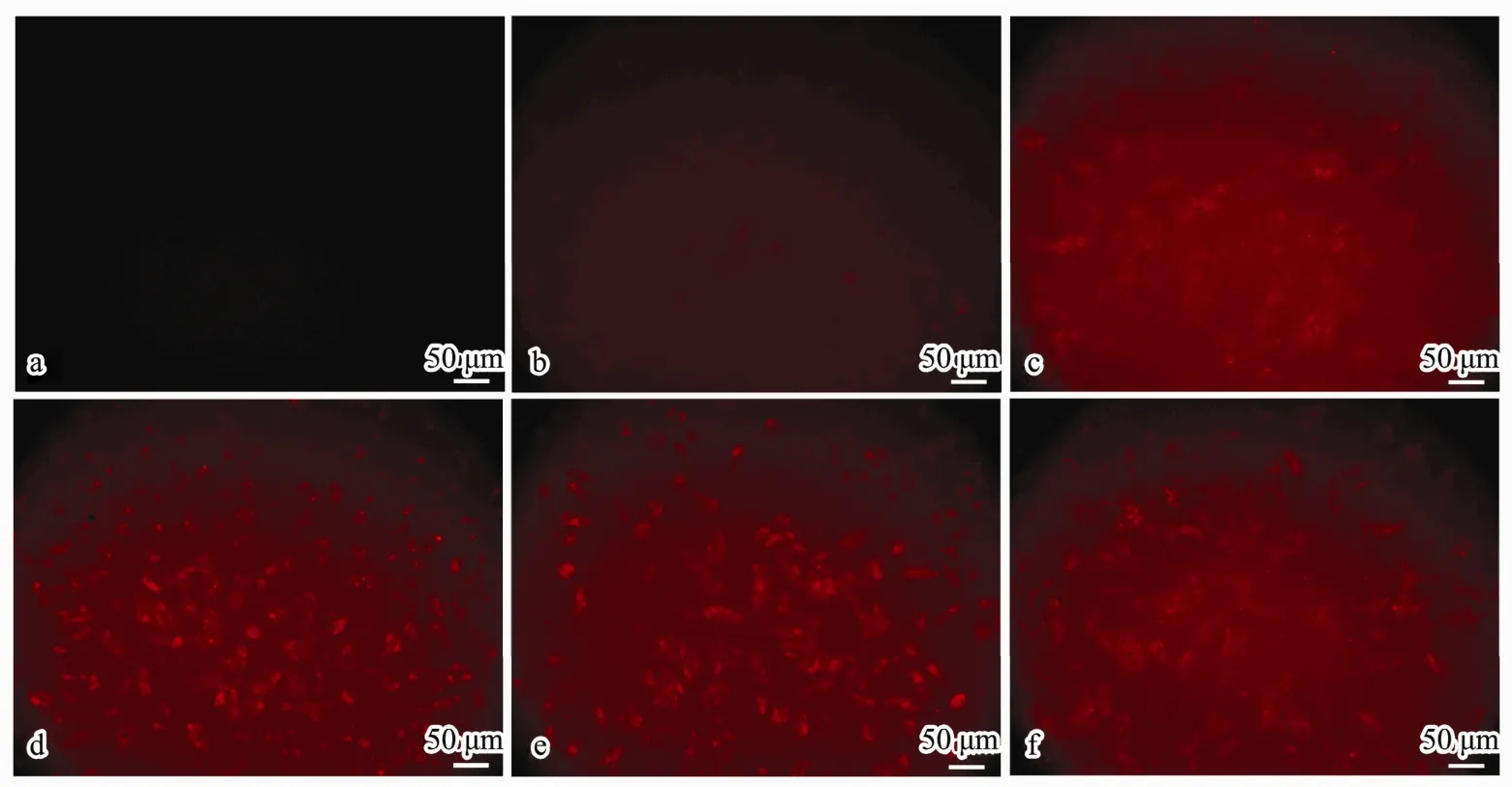
Fig.7 Fluorescent images of siRNA transfection in A549 cells observed using fluorescence microscope after treatment with CSsNPs loaded with different amounts (0.375, 0.75, 1.5, and 3 OD) of Bcl2-siRNA-Cy3 (b-e) compared to those for the normal (a) and positive (f) controls (200×).
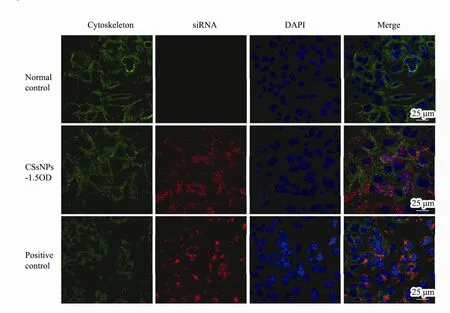
Fig.8 Images of CSsNPs loaded with 1.5 OD siRNA in A549 cells were observed by laser scanning confocal microscopy and compared to those for the normal and positive controls. The cytoskeleton was stained with actin (green fluorescence),and nuclei were stained with DAPI (blue fluorescence).
4 Discussion
Nanoparticles (NPs) are proven to be effective tools for the delivery of nucleic acids without side effects (Tahamtan et al., 2015; Hosseini et al., 2016), because NPs based on chitosan can be formed by ionic gelation, and can bind with negatively charged nucleic acids. However, the size of the nanoparticle is important, because smaller sized NPs can escape detection by phagocytes in blood, which makes them viable for wider use in drug and gene vector in vivo (Jadidi-Niaragh et al., 2016). However, there are few reports on the kind of gene products that are suitable for delivery by small-sized nanoparticles.
In this study, we compared the size, PDI, and zeta potential of CSNPs prepared with different concentrations of chitosan (MW 160 kDa) and TPP. When the ratio of CS to TPP (1 mg mL-1) was 10:1 (w/w), the CSNPs had a smaller size (100-200 nm), lower PDI (below 0.2), and more stable zeta potential, which indicates that they were monodisperse and maintained a stronger penetration capacity (Table 1). Additionally, the images observed by TEM confirmed that the CSNPs of about 100-200 nm were uniform in size (Fig.1). When NPs encapsulated plasmids or siRNA, there were no significant changes to their morphological characteristics (Fig.4, Table 2).
Cytotoxicity and histocompatibility are very important for medical materials, and there are corresponding ISO standards. Accordingly, the cytotoxicity of CSNPs was evaluated by MTT assay, which demonstrated that the CSNPs prepared in this study had low toxic effects on L929 cells during 72 h of treatment (Fig.2). Further, the biodegradability and histocompatibility of medical materials in vivo were assessed in a mouse model via intramuscular implantation. It has been reported that when a biomaterial is implanted, the local tissue reacts initially to the injury, and then to the presence of the material. Inflammation is the most common reaction to all injury forms (Liang et al., 2011). To evaluate the biocompatibility of CSNPs, the slides for histological examination were stained with hematoxylin and eosin (H&E) and observed under a microscope. It seems that the nanoparticles do not cause obvious tissue damage or inflammation at 1 w, 2 w,and 3 w after treatment (Fig.3), which might be because of the smaller size and better dispersibility. In conclusion,nanoparticles prepared in this study had a low cytotoxicity and good tissue biocompatibility.
To investigate the loading capacity for nucleic acids,most studies use UV spectrophotometry or agarose gel electrophoresis assays. In this study, we used these two methods to detect the gene-loading capacity of CSNPs. It seems that CSNPs have a high loading efficiency for both plasmids and siRNA (Fig.5, Table 2). However, the cell transfection of GFP-plasmid or siRNA-Cy3 to A549 cells by CSNP vector was detected differently by fluorescence microscopy and laser scanning confocal microscopy(Figs.6-8).
Lip3000 and HiPerFect were used for the positive controls, because they are recognized as the most effective transfection reagents for plasmids and siRNA respectively. CSNPs can bind with nucleotide sequences and transfer them to cancer cells, and the transfection efficiency was gradually increased as the encapsulation amounts increased. However, for plasmids with a sequence of 7000 bp, the transfection efficiency of CSpNPs loaded with 14-140 μg plasmids was much lower than the positive control (Fig.6). Additionally, in the siRNA-Cy3(sequence of about 20 bp) encapsulation groups, the fluorescence could be enhanced with increased loading amounts of CSsNPs (Fig.7), and the transfection rate of the CSsNPs loaded with 1.5 OD siRNA was higher than that of the positive control (Fig.8). Therefore, CSNPs might be more suitable as siRNA vectors than plasmid vectors, because the former has a much higher transfection efficiency, which might be related to the length of the sequence. These results imply that CSNPs could be potentially applied as safe and efficient delivery systems for nucleic acids with short sequences.
5 Conclusion
In this study, CS (MW 160 kDa) was chosen to prepare CSNPs successfully by the ionic gelation method, which produced CSNPs with smaller particle size, stable dispersibility, low cytotoxicity, good tissue biocompatibility,and high gene-loading efficiency. CSNPs could transfer the plasmids or siRNAs to cells. However, the CSsNPs had a significantly higher transfection efficiency than CSpNPs. Therefore, our results imply that CSNPs might be more suitable as safe and efficient vectors for siRNA rather than for plasmids.
Acknowledgements
This work was supported by the Natural Science Foundation of Shandong Province (No. ZR2014HP011),Qingdao Young Scientist Applied Basic Research Fund(No. 15-9-1-51-jch), Youth Foundation of The Affiliated Hospital of Qingdao University (No. 2417), and the National Natural Science Foundation of China (No. 8140 1899).
 Journal of Ocean University of China2018年5期
Journal of Ocean University of China2018年5期
- Journal of Ocean University of China的其它文章
- Effect of Different Dietary Protein and Lipid Levels on the Growth, Body Composition, and Intestinal Digestive Enzyme Activities of Juvenile Yellow Drum Nibea albiflora (Richardson)
- Methylation Status of the Follistatin Gene at Different Development Stages of Japanese Flounder(Paralichthys olivaceus)
- Taxonomic Clarification of A Well-Known Pathogenic Scuticociliate, Miamiensis avidus Thompson &Moewus, 1964 (Ciliophora, Scuticociliatia)
- Structural Variation Analysis of Mutated Nannochloropsis oceanica Caused by Zeocin Through Genome Re-Sequencing
- A Comparative Study on Hydrodynamic Performance of Double Deflector Rectangular Cambered Otter Board
- Characterization of Polysaccharides Extracted from a Cultivated Brown Alga Costaria costata During the Harvest Period
