GmDRR1, a dirigent protein resistant to Phytophthora sojae in Glycine max (L.) Merr.
CHEN Qing-shan, YU Guo-long, ZOU Jia-nan, WANG Jing, QIU Hong-mei, ZHU Rong-sheng,CHANG Hui-lin, JIANG Hong-wei, HU Zhen-bang, LI Chang-yu, ZHANG Yan-jiao, WANG Jin-hui,WANG Xue-ding, GAO Shan, LIU Chun-yan, QI Zhao-ming, FU Yong-fu, XIN Da-wei
1 Key Laboratory of Soybean Biology, Ministry of Education/College of Agriculture, Northeast Agricultural University, Harbin 150030, P.R.China
2 Key Laboratory of Soybean Biology (Beijing), Ministry of Agriculture/National Key Facility of Crop Gene Resource and Genetic Improvement/Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, P.R.China
1. Introduction
Dirigent (DIR) proteins are a class of proteins which dictate the stereochemistry of a compound synthesized by other enzymes. The term “dirigent” originates from the Latin word “dirigere” (to align or guide) (Hosmaniet al.2013; Liet al. 2014). DIR proteins were reported playing an important role in both the correct patterning of lignin deposition and response to pathogen infection and abiotic stress in plants (Ganget al. 1999; Davin and Lewis 2000;Burlat 2001; Behret al. 2015). Hundreds of dirigent and dirigent-like proteins have been discovered in vascular plants, such asArabidopsis, rice, spruce, wheat, cotton,andBoea hygrometrica(Ralphet al. 2006, 2007; Ma and Liu 2015). DIR proteins could be subdivided into five groups: DIR-a, DIR-b, DIR-c, DIR-d, and DIR-e. Due to an increasing number of DIR/DIR-like proteins, DIR-b and DIR-d subfamilies are combined together as DIR-b/d family(Ralphet al. 2007). BhDIR1 is an earlier identified DIR protein inB.hygrometrica, which is involved in response to dehydration status, phytohormones, signaling molecules and temperature stresses (Wuet al. 2009). A transcription factor AtMYB14 is capable of activating the synthesis and export of BhDIR1 (Kosmaet al. 2014). Enhanced Suberin 1(ESB1) plays an essential role in the correct formation of Casparian strips inArabidopsis(Hosmaniet al. 2013). In wheat, the TaDIR13, belonging to the DIR-b/d subgroup, is mainly both participated in regulating lignan biosynthesis and responsible for the resistance toPhytophthorasojae(Ma and Liu 2015).
P.sojaeKaufm. & Gerd., detected firstly in the United States in 1948, induces phytophthora root and stem rot(PRSR) in soybean (Kaufmann and Gerdemann 1958;Dorranceet al. 2007).P.sojaecould survive in soil for several years and create destructive damage to soybean yield in high moisture conditions (Dorranceet al. 2007;Tianet al. 2016). In 1991, PRSR was first described in Heilongjiang Province, a main soybean production area in China (Shenet al. 1991). Million tons of yield loss were caused by PRSR in the past decade in the world (Koenning and Wrather 2010; Pinget al. 2016). Many methods had been applied to control this disease, such as using resistant cultivars, seed coating and reducing the soil moisture (Pinget al. 2016). Genetic data show that resistance in soybean is controlled by interaction on a gene-for-gene basis with theP.sojae. To date, 19Rps(resistance toPhytophthora sojae) genes plus 25 alleles have been confirmed to be related to PRSR. These genes/alleles areRps1(1a, 1b,1c, 1d, and 1k),Rps2,Rps3(3a, 3b, and 3c),Rps4,Rps5,Rps6,Rps7,Rps8,Rps9,Rps10,RpsUN1,RpsYu25,RpsYD29,RpsJS,RpsYB30,RpsZS18,RpsSu,RpsYD29,and an unnamedRpsgene reported in Japanese cultivar Wasehiroge (Demirbaset al. 2001; Wenget al. 2001;Burnhamet al. 2003; Grauet al. 2004; Gaoet al. 2005;Gordonet al. 2006; Fanet al. 2009; Yaoet al. 2010;Sugimotoet al. 2011; Sunet al. 2011; Wuet al. 2011; Linet al. 2013; Zhanget al. 2013). No DIR genes in soybean had been reported to be involved in the resistance toP.sojaenow. Here, we performed 2-DE analysis combined with MALDI-TOF-MS for protein profiles in soybean leaves responding toP.sojae. And we identified a DIR protein GmDRR1 (Glycine maxDisease Resistance Response 1)showing resistance toP.sojae.
2. Materials and methods
2.1. Plant materials and stress treatments
Eleven resistant and 11 susceptible germplasm varieties were used in this study (Table 1). The seeds were sterilized by chlorine gas at fume hood for 16 h and placed on vermiculite:water (1:2, w/v) and covered with the same wet vermiculite, seeds were germinated in the dark for 4 days at 25°C. Plants were grown in a growth chamber at 25°C and 70% humidity in 16 h light/8 h dark cycles for 4 weeks. Soybean hypocotyls were inoculated with mycelia ofP.sojaeat the position of 2–3 cm below the cotyledon nodes in dark for 12 h, then transfer to 16 h light/8 h dark cycles (Donget al. 2011; Wanget al. 2011). For the 2-DE,the unifoliolate leaves were harvested at 12, 24, 36, and 48 h after inoculation withP.sojaerace 1 and frozen in liquid nitrogen and stored at –80°C. For qRT-PCR, the first trifoliolates, stems, and roots were sampled at 0, 6, 12, 24,and 48 h after infection withP.sojaerace 1.
2.2. Protein extraction and 2-DE
Each sample was conducted for three times. Twenty hypocotyls of soybean seedlings were pooled. Total proteins from soybean unifololiates were extracted using acetone/TCA propitiation method according the Bio-Rad 2-D manual.At least 1.5 mg proteins were loaded to a nonlinear gradient IPG strip (24 cm, pH 4–7, Bio-Rad, America). 2-DE was performed in a horizontal apparatus (IPGphor3 from GE Healthcare Life Science and Hoefer 600 SE from Amersham Pharmacia Biotech, America) (Linket al. 1999; Vanrobaeyset al. 2005; Roccoet al. 2013). The different spots wereanalyzed by the software of Image MasterTM 2D Platinum 6.0. And the SDS-PAGE gel was stained using colloidal Coomassie blue. The analysis of MALDI-TOF-MS was completed by the corporation of Sangon Biotech (Shanghai,China) Co., Ltd.
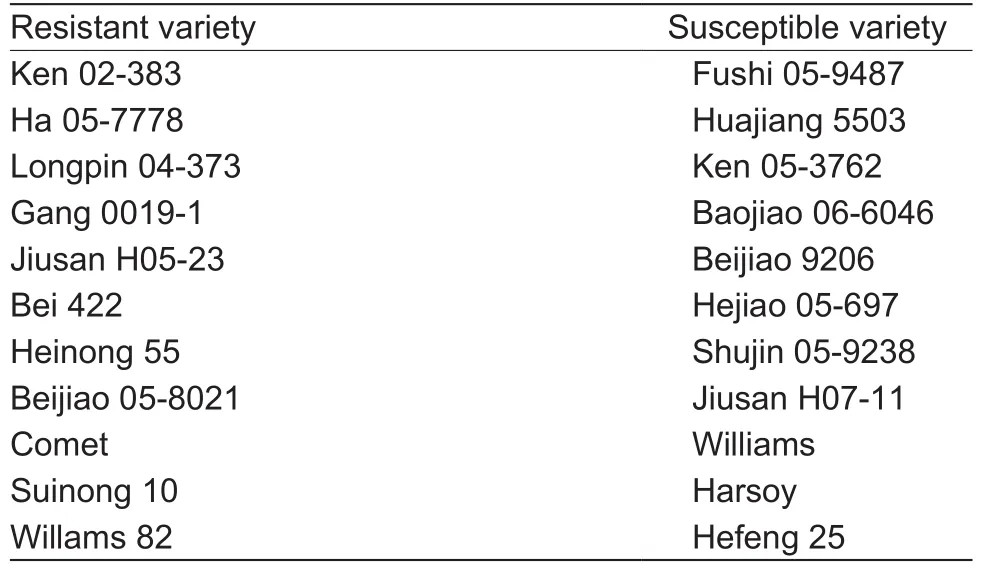
Table 1 The name of 11 resistant-soybean and 11 susceptiblesoybean germplasms
2.3. Isolation of GmDRR1 sequence
Depending on the result of 2D-MS, the full coding sequence of GmDRR1 was amplified from soybean Suinong 10 by RT-PCR using the primers ofGmDRR1-F andGmDRR1-R(GenBank accession no. HM143918.1:1..765, NCBI protein no. ADG86640.1) (Table 2). Primers were designed according to the reference genome of Williams 82 in Phytozome database (http://www.phytozyme.net/). The thermal cycler was programmed with an initial 5 min at 95°C,followed by 35 cycles of 30 s at 95°C, 30 s at 60°C and an extension of 1 min at 72°C, and with a final extension at 72°C for 10 min. Then theGmDRR1PCR product was purified by a gel recycle and cloned into the GATEWAY donor vector pGWC (Chenet al. 2006), andGmDRR1gene was transferred into a binary vector pLeela by an LR reaction(Invitrogen, America).
2.4. Expression of GmDRR1 in Nicotiana bentha miana
Plasmids (pLeela-GmDRR1) and the empty vector pLeela were transformed intoAgrobacterium tumefaciensstrain EH105 by heat shock, respectively. Leaves from 4-wk-oldN. benthamianaplants were infiltrated withAgrobacteriumbacteria (OD600≈ 0.5) in infiltration buffer (10 mmol L–1MgCl2,10 mmol L–1MES, pH 5.6). A image processing software can use to measure area. Soybean plants were grown with a photoperiod of 16 h light/8 h dark and maintained at 22°C and 70% relative humidity in a greenhouse.
2.5. Quantitative reverse transcription (qRT-PCR)
Total RNA was extracted using the NucleoSpin RNA Plant Extraction Kit (Machery-Nagel, Germany). For analysis of gene expression, the first-strand cDNA was synthesized from 1 μg of RNA using AMV (avian myeloblastosis virus reverse transcriptase) reverse transcriptase (Promega) and an oligo (dT) primer (Microsynth, Switzerland), according to the manufacturer’s instructions. For quantitative PCR,5 μL of 1/100 dilution of cDNA were combined with SYBR Master Mix. PCRs were performed in triplicates with the Real Time PCR System (7500, Applied Biosystems).Data were collected and analyzed with the ABI (Applied Biosystems and Invitrogen) Analyzing Program. The soybean housekeeping geneUKN1(Glyma12g02310)was used as the internal control. Each qRT-PCR was run in three biological replicates. Data derived from three biological repeats were statistically analyzed by ANOVA(one-way ANOVA) consideringP≤0.05 as significantly different.

Table 2 Primer list in this study
2.6. Promoter element analysis
TheGmDRR1promoters, named asGmDRR1Hefeng25p,GmDRR1Suinong10p,GmDRR1williamsp, andGmDRR1williams82p,were amplified, respectively, from the genome of Hefeng 25, Suinong 10, Williams, and Williams 82 based on Phytozome database (http://www.phytozome.net) with the primersGmDRR1p-F andGmDRR1p-R (Table 2).The sequences were cloned into the Fu39-2 vector(Wanget al. 2013) by LR reaction (Invitrogen, China),resulting in plasmid Fu39-2-GmDRR1Hefeng25p, Fu39-2-GmDRR1Suinong10p, Fu39-2-GmDRR1williamsp, and Fu39-2-GmDRR1williams82p, respectively.
2.7. Promoter haplotype analysis in soybean germplasm
We target the 3 kb region upstream of GmDRR1 start codon as the promoter region. The genomic sequence of this region in 302 soybean germplasms (Zhouet al. 2015)were used to identify the haplotype. The haplotype blocks were detected by the Haploview software (Barrettet al.2005). Then some modification and change were based on the Perl script.
2.8. Confocal microscopy
The coding sequence of GmDRR1 was transferred into vector Fu39-2 (Wanget al. 2013). TheGmDRR1-GFP(green fluorescent protein) expression level inArabidopsisprotoplast was detected as known methods (Bessereret al.2012; Yanget al. 2014; Thazar-Poulotet al. 2015). 4-wk-old seedlings ofArabidopsiswere used for protoplast isolation.The protoplasts were derived from the leaf mesophyll cells ofArabidopsis. Transfected cells were imaged using a TCS SP2 Confocal Spectral Microscope Imaging System(Leica, Germany). Excitation/emission wavelengths of 488/500–530 nm were used for GFP.
2.9. Bioinformatic analysis of protein spots
All spectra of proteins were subjected to database searching against NCBInr, SwissProt, and MSDB databases using the online Mascot Program (http://www.matrixscience.com).The searching parameters were as follows: a mass tolerance of 30 ppm for peptides, 0.5 Da for fragments, and maximally one missed cleavage site. Only significant hits, as defined by the Mascot probability analysis (P<0.05), were accepted.Analysis of GmDRR1 protein structure was performed by SMART (http://smart.embl-heidelberg.de/). Sequence alignments were performed using ClustalX 2.0 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The phylogenetic analysis of GmDRR1 and various heterologous DIR members was performed using MEGA5.1 software by the Maximum Likelihood (Tamuraet al. 2011).
3. Results
3.1. Two-dimensional gel electrophoresis (2-DE) and biological mass spectrometry
Nineteen differential protein spots were found by the comparative 2-DE analysis, including 10 upregulating spots, 8 downregulating spots, and 1 specific spot (spot 10) in the sample inoculated withP.sojae(Fig. 1-A). Eight upregulating-spots, 1 downregulating-spot (spot 9), and spot 10 were further analyzed by the MALDI-TOF-MS (Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry, Bruker Daltonics, Italy). Finally, 8 candidate proteins were confirmed in database of NCBhr, SwissProt,and MSDB (Table 3). Spot 3 is an F-box protein containing the leucine-rich repeat (LRR) domain and F Box domain(FBD). FBD protein has the transcriptional factor function and is a disease response protein involved in jasmonic acid (JA) induced defense response (Xuet al. 2009).Spot 4 is a kinase receptor, which might be involved in the signal transduction in biotic and abiotic stresses (Zipfelet al. 2014). Spot 5 is a protein containing Zeta-3 subunit,which might be involved in the regulation of the coatomer assembly (Stephenset al. 2000). Spot 6 is a RuBisCO precursor and spot 7 is a RuBisCO large chain. Spot 8 belongs to the antioxidant super protein family containing a Peroxiredoxin-2E-2 domain. Spot 9 is a Cys peroxiredoxin,whose N-terminus has a conserved Cys residue. Spot 10 is a dirigent protein 21-like. Because dirigent proteins are involved in the disease resistance in soybean (Liet al. 2014),we named this dirigent protein 21-like as GmDRR1.

Fig. 1 Comparison profile of proteins from Suinong 10 leaves inoculate with Phytophthora sojae race 1. A, proteins mixture of the unifoliolate leaves were harvested at 12, 24, 36, and 48 h after inoculation with P. sojae race 1. B, proteins mixture of the unifoliolate leaves were harvested at 12, 24, 36, and 48 h after mock inoculated (wounded) without the pathogen. pI, isoelectric point.
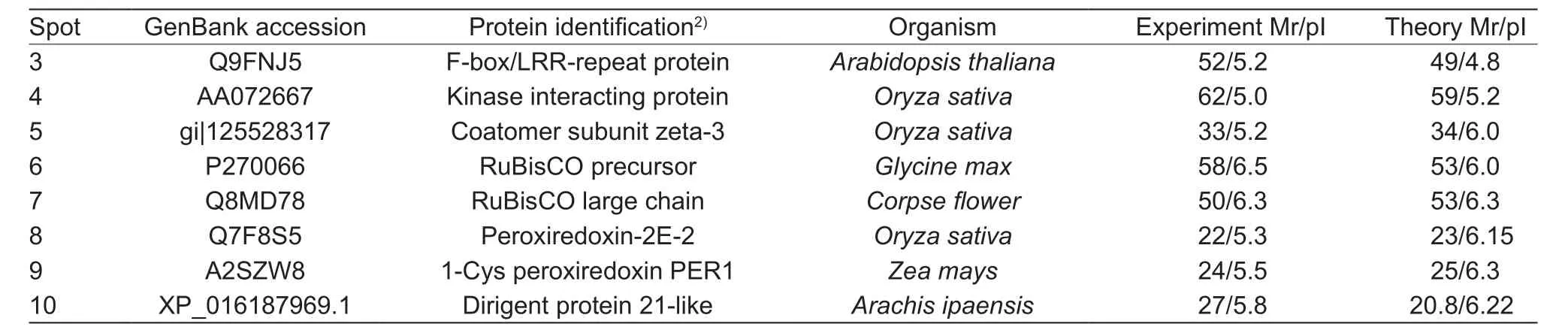
Table 3 List of identification proteins by MADI-TOF-MS (Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry)1)
3.2. Orthology of GmDRR1 protein family
TheGmDRR1coding sequence is 765 bp in length. A conserved DIR domain is located in the region from aa106to aa253. This gene is located in the B1 linkage group in the interval of molecular markers Satt597 and Sct026(Fig. 2-A), indicating its disease-resistant relationship. The phylogenetic tree analysis revealed thatGmDRR1was belong to the DIR-b/d family. We found 50 genes similar toGmDRR1in soybean genomeby alignment analysis.Among them, 27 of 50 belong to the DIR-b family, 10 belong to DIR-c subfamily, 4 belong to DIR-d subfamily, 9 belong to DIR-e subfamily, respectively (Fig. 2-B). In general,DIR-a and DIR-e mainly appear in monocotyledon plants.DIR-c could be further divided into 2 sub-families: one belongs to monocotyledon plants and the other belongs to dicotyledonous plants. By the alignment analysis of GmDRR1 proteins in 11 susceptible and resistant soybean germplasm, we found that there was no significant difference in the amino acid sequence, suggesting the difference in susceptible and resistant varieties may not be due to GmDRR1 protein sequences (Appendix A). It may be due to the differences in promoter.
3.3. GmDRR1 may targete on the plant cytomembrane
For the subcellular localization analysis of GmDRR1 proteins, theGmDRR1gene fused to fluorescent tag (GFP)was expressed in theArabidopsisprotoplasts. Observations with confocal microscope demonstrated that GmDRR1-GFP fluorescence maybe localized on the cytomembrane ofArabidopsisprotoplast (Fig. 3). This support that GmDRR1 may be a cytomembrane located protein.
3.4. GmDRR1 inhibited P. sojae in N. benthamiana
To examine whether GmDRR1 was involved in the disease resistance in soybean. The plasmid pLeela-GmDRR1and zoospore ofP.sojaerace 1 were co-infiltrated into the leaves ofN.benthamiana.The disease lesions diameter of the plasmid pLeela-GmDRR1and zoospore ofP.sojaerace 1 was about 4-fold than the plasmid pLeela-hGFPand zoospore ofP.sojaerace 1 (Fig. 4), suggesting GmDRR1 significantly inhibitedP.sojaeinfection.
3.5. Transcription characters of GmDDR1 in various soybean varieties
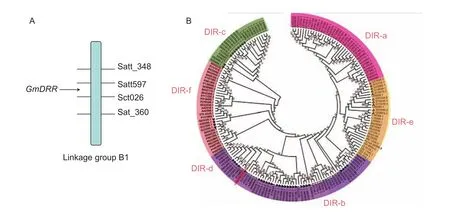
Fig. 2 Location and phylogenetic tree of Glycine max Disease Resistance Response 1 (GmDRR1). Amino acids of 179 dirigent or dirigent-like (DIR) proteins are analyzed by the Maximum Likelihood (ML) using MEGA 5.1 (–ln=2 880.02, model: WAG+F).Subfamilies DIR-a, DIR-b/d, DIR c, DIR-e, and DIR-f are indicated by different colours. The proteins belong to Glycine max,Arabidopsis thaliana, Solanum lycopersicum, Populus trichocarpa, Oryza sativa, Zea mays, Sorghum bicolor, and Physcomitrella patensV1.6.

Fig. 3 Subcellular localization of GmDRR1 (Glycine max Disease Resistance Response 1) proteins. Subcellular localization was investigated in Arabidopsis protoplasts under a confocal microscope. The fluorescent distribution of humanized green fluorescent protein (hGFP) and the fusion protein GmDRR1-hGFP were observed under white light, UV light, and red light, respectively.
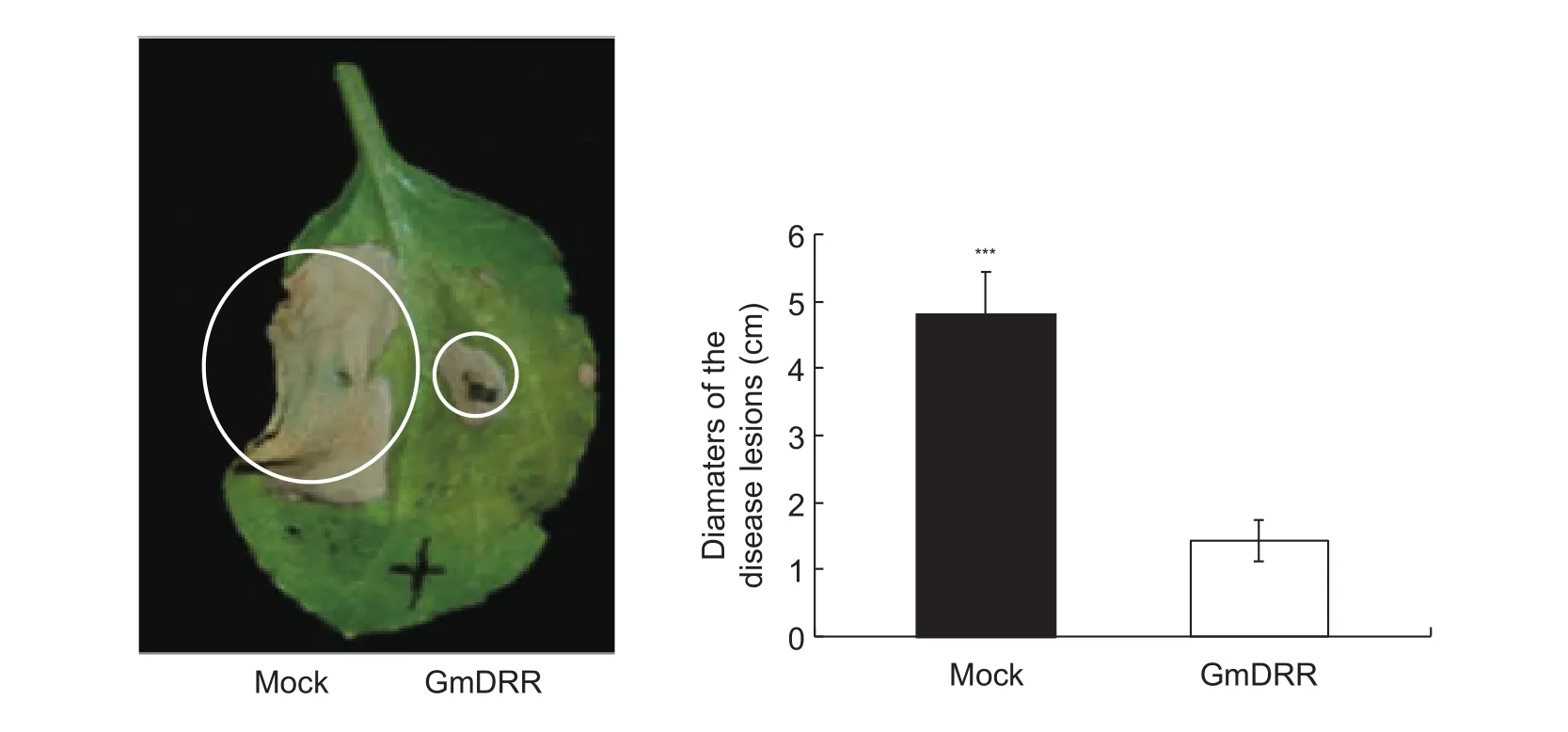
Fig. 4 GmDRR1 (Glycine max Disease Resistance Response 1) inhibited the cell death induced by Phytophthora sojae in tobacco.The left part were infiltrated with P. sojae zoospores and Agrobacterium tumefaciens carrying pLeela-hGFP (humanized green fluorescent protein), the right part were infiltrated with both P. sojae zoospores and Agrobacterium tumefaciens carrying pLeela-GmDRR. Bars are SD (n=5), and asterisks denote significant differences (***, P<0.001, one-way ANOVA). These experiments were repeated three times with similar results.

Fig. 5 Expression analyses of GmDRR1 (Glycine max Disease Resistance Response 1) in stems, roots, and leaves at different times after inoculation of Phytophthora sojae race 1 by qRT-PCR. Cultivar Suinong 10 is resistant to P. sojae race 1, while Hefeng 25 is susceptible to P. sojae race 1. Bars mean SD.
Soybean varieties Suinong 10 and Hefeng 25 were inoculated byP.sojaerace 1, and then the expression pattern ofGmDRR1in stems, leaves, and roots was analyzed with qRT-PCR (Fig. 5). In the leaf samples, a significant peak was recognized in Suinong 10 but not Hefeng 25 at 6 hours post inoculation (hpi) race 1, while there seems no reaction forGmDRR1of Hefeng 25 in leaf samples (Fig. 5-C). In the stem sample,GmDRR1expression in Suinong 10 began increasing at 6 hpi and reached a peak at 12 hpi, followed by a decline to 48 hpi. In Hefeng 25,GmDRR1expression increased slowly and reached a small peak at 6 hpi, then declined to a base level from 12 to 48 hpi (Fig. 5-A). In the root sample, the level ofGmDRR1transcripts in Suinong 10 increased slowly and reached a peak at 12 hpi, and then decreased and reached a zero level at 24 hpi. In Hefeng 25,GmDRR1transcripts accumulated and quickly reached a peak at 6 hpi, and then declined slowly to zero level at 24 hpi(Fig. 5-B). The results indicated that the difference ofGmDRR1expression in susceptible and resistant varieties mainly existing in stems and leaves.
3.6. Promoter elements analysis
The upstream 2.5 kb sequence ofGmDRR1coding sequence was cloned into the vector Fu76 (Wanget al.2013) and confirmed by sequencing. By the analysis of PlantCARE software (Lescotet al. 2002), some motifs related to stress and pathogen resistant were found,including MBS sites, MYB binding sites, TC-rich repeats,and HSE motif (Fig. 6). The difference of promoters between resistant and susceptible varieties could be found,suggesting that the promoters may confer to the difference ofGmDRR1gene transcription and be involved inP.sojaeresistance.
3.7. Haplotype diversity in GmDRR1 promoter region
To further understand GmDRR1 promoter characters related to the gene expression, the promoter region of GmDRR1 in 302 soybean germplasm genomes were aligned. Finally,44 SNPs (single nucleotide polymorphisms) were found.This haplotype could be divided into 77 subtypes (Fig. 7-A).Among them, H8 haplotype was existing in most resistant varieties, while H77 was existing in most susceptible varieties. We aligned the sequence of H8 with H77 and 7 SNPs were revealed (Fig. 7-B), including C to A, G to C, C to A, T to A, T to C, T to C, and T to A (Appendix B).Additionally, two single nucleotide insertions (T and A) were found in the H77 haplotype. The difference might be a key factor regulating the GmDRR1 transcription response to theP.sojaeinduction.
4. Discussion
4.1. GmDRR involved in the P. sojae resistant of soybean
To investigate the changes of protein profiles in soybean infected withP.sojae, we carried out 2-DE analysis of the total proteins in soybean leaves from the mixture of 3-fold points after inoculated withP.sojae. Even though there were similar protein profiles displayed betweenP.sojae-infected and mock samples, variations in quality and abundance of special protein spots were significant. As little information on proteomic responses of soybean occurring in host during infection byP.sojae, some candidate protein spots might not be real proteins involved in disease resistance. At last, we selected GmDRR1 as the target protein for further analysis,and our results made us to believe that GmDRR1 was one of proteins resistant toP.sojaein soybean.
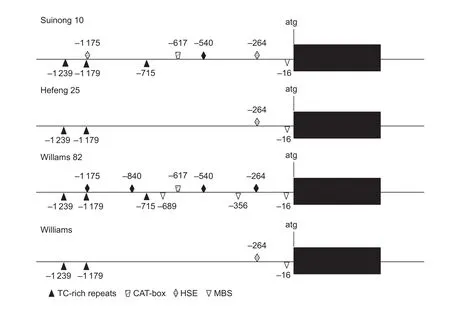
Fig. 6 Analysis of cis-acting regulatory element in GmDRR1 (Glycine max Disease Resistance Response 1) promoter between resistant and susceptible varieties.
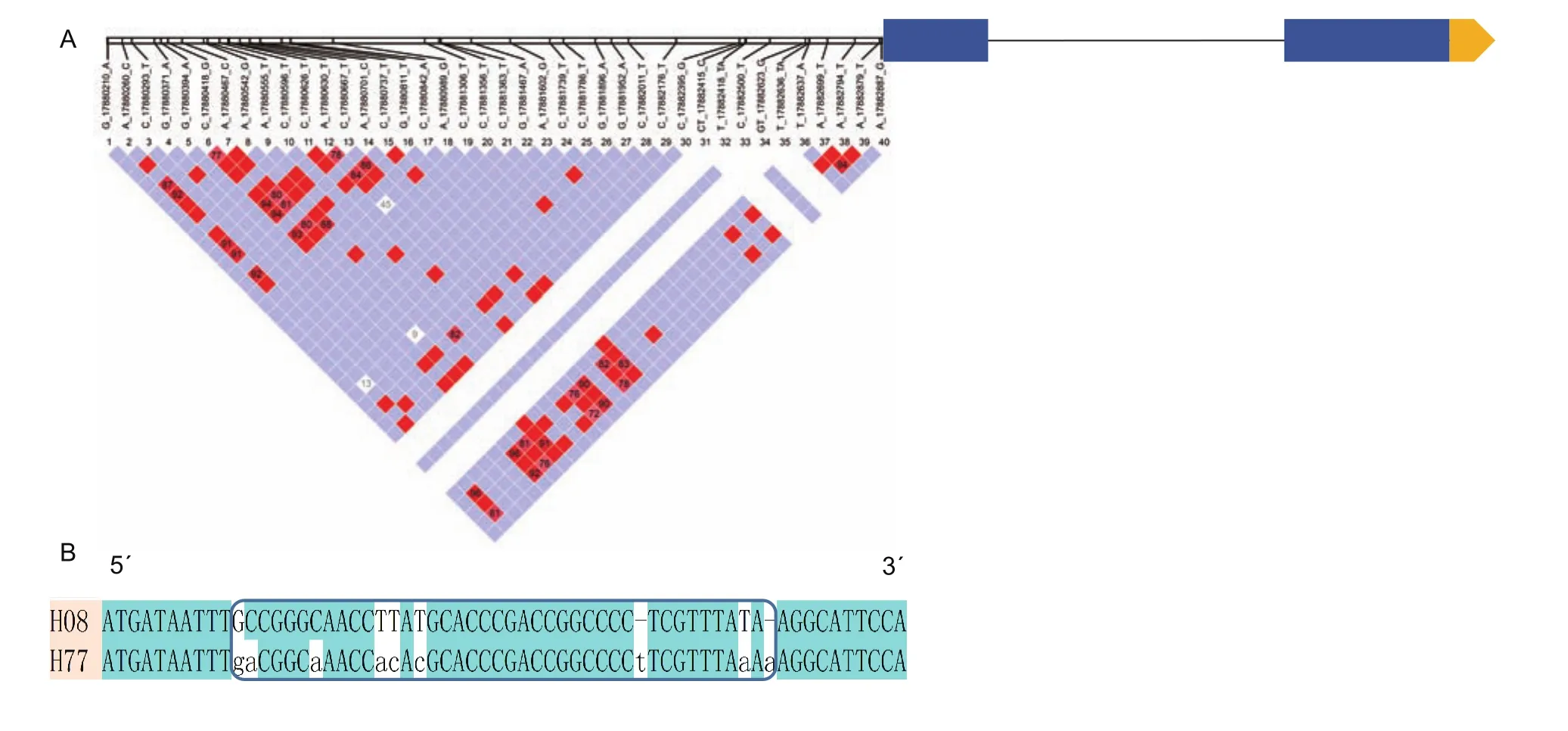
Fig. 7 GmDRR (Glycine max Disease Resistance Response) gene structure and haplotype in 302 soybean germplasms. A, the SNP (single nucleotide polymorphisms) distribution in promoter region. B, the comparation of H8 and H77 haplotypes.
DIR and DIR-like proteins had been found in all of the major terrestrial plants (Davin and Lewis 2000) and are involved in the production of both lignin and lignan in adaptation of aquatic plants to environment (Weidenbachet al. 2016). Here, GmDRR1 was a DIR protein showing the resistance toP.sojaein soybean, so the regulation strategy ofGmDRR1related to resistance might be depend on the lignin synthesis. It had been confirmed that the lignin synthesis is often induced at the site of pathogen attack and participated in the plant defense mechanism (Weidenbachet al. 2016).
4.2. Promoter might play a pivotal role in regulate GmDRR transcription
No significant difference was found in the coding sequence ofGmDRR1in 11 resistant-soybean or 11 susceptible-soybean varieties. Only several single amino acid differences were found, and theses polymorphisms were not conserved and not related to resistant or susceptible varieties, indicating that the resistant strategy ofGmDRR1might not be due to the difference of coding regions. In the promoter region of Suinong 10, Hefeng 25, Williams 82, and Williams, we found that the number of regulating elements is different between resistant and susceptible soybean cultivars, including some disease related elements, such as MBS, MYB, and TC-rich repeats (Xuet al. 2010). The number of TATA- and CAAT-boxes were 122 and 82 in resistant cultivars Williams 82 and Suinong 10, respectively, whereas in susceptible cultivars Hefeng 25 and Williams, the number is 78 and 80, respectively. The number difference of MBS, MYB and TC-rich repeats in resistant and susceptible cultivars is not significant. Therefore, the transcription regulation ofGmDRR1gene may be more important than protein difference for soybean resistant toP.sojae. In the same time, the promoter region existing multifarious haplotypes,and H8 and H77 are the mainly-different haplotypes between resistant and susceptible varieties. And the SNPs existing in the promoter region might control the transcription ofGmDRR1gene in response toP.sojae.However, more work is needed to detect the strategy ofGmDRR1regulating pathogen resistance.
5. Conclusion
GmDRR1 as the different protein pot which is identified by MS between susceptible and resistant soybean varieties.We analyzed the gene transcription, location, and promoter element character ofGmDRR1. Results support thatGmDRR1is a disease resistant gene response toP. sojaeinfection. The results of tobacco infiltration support that GmDRR directly involved in theP. sojaeresistant.
Acknowledgements
This study was financially supported by the Academic Skeleton Support Plan of Department of Education of Heilongjiang Province, China (1254G011), the National Natural Science Foundation of China (31271747, 31471516,31400074, 31401465, 31501332), the National High-Tech R&D Program of China (the 863 Program, 2013AA102602),the Research Fund for the Doctoral Program of Higher Education of China (20122325120015), the Academic Backbone Project of Northeast Agricultural University, China(15XG02), and the Talented Young Project of Northeast Agricultural University, China (518062).
Appendicesassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Barrett J C, Fry B, Maller J, Daly M J. 2005. Haploview: Analysis and visualization of LD and haplotype maps.Bioinformatics,21, 263–265.
Behr M, Legay S, Hausman J F, Guerriero G. 2015. Analysis of cell wall-related genes in organs ofMedicago sativaL.under different abiotic stresses.International Journal of Molecular Sciences,16, 16104–16124.
Besserer A, Burnotte E, Bienert G P, Chevalier A S, Errachid A, Grefen C, Blatt M R, Chaumont F. 2012. Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121.The Plant Cell,24,3463–3481.
Burlat V, Kwon M, Davin L B, Lewis N G. 2001. Dirigent proteins and dirigent sites in lignifying tissues.Phytochemistry,57,883–897.
Burnham K D, Dorrance A E, Francis D M, Fioritto R J, Martin S K S. 2003. Rps8, a new locus in soybean for resistance toPhytophthora sojae.Crop Science,43, 101–105.
Chen Q J, Zhou H M, Chen J, Wang X C. 2006. Using a modified TA cloning method to create entry clones.Analytical Biochemistry,358, 120–125.
Davin L B, Lewis N G. 2000. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis.Plant Physiology,123, 453–462.
Demirbas A, Rector B, Lohnes D, Fioritto R, Graef G, Cregan P, Shoemaker R, Specht J. 2001. Simple sequence repeat markers linked to the soybean genes forPhytophthoraresistance.Crop Science,41, 1220–1227.
Dong S, Yin W, Kong G, Yang X, Qutob D, Chen Q, Kale S D, Sui Y, Zhang Z, Dou D. 2011.Phytophthora sojaeavirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity.PLoS Pathogens,7, e1002353.
Dorrance A E, Mills D, Robertson A E, Draper M A, Giesler L,Tenuta A. 2007. Phytophthora root and stem rot of soybean.The Plant Health Instructor,1, doi: 10.1094/PHP-2004-0309-01-RS
Fan A Y, Xiaoming W, Fang X P, Wu X F, Zhu Z D. 2009.Molecular identification of phytophthora resistance gene in soybean cultivar Yudou 25.Acta Agronomic Sinica,35,1844–1850. (in Chinese)
Gang D R, Costa M A, Fujita M, Dinkova-Kostova A T, Wang H B, Burlat V, Martin W, Sarkanen S, Davin L B, Lewis N G. 1999. Regiochemical control of monolignol radical coupling: a new paradigm for lignin and lignan biosynthesis.Chemistry & Biology,6, 143–151.
Gao H, Narayanan N N, Ellison L, Bhattacharyya M K. 2005.Two classes of highly similar coiled coil-nucleotide bindingleucine rich repeat genes isolated from the Rps1-k locus encodePhytophthoraresistance in soybean.Molecular Plant-Microbe Interactions,18, 1035–1045.
Gordon S G, St Martin S K, Dorrance A E. 2006. Rps8 maps to a resistance gene rich region on soybean molecular linkage group F.Crop Science,46, 168–173.
Grau C R, Dorrance A E, Bond J, Russin J. 2004. Fungal diseases. In: Boerma H R, Specht J E, eds.,Soybeans:Improvement, Production and Uses. 3rd ed. Agronomy Monograph. American Society Agronomy. Madison, WI.pp. 679–763.
Hosmani P S, Kamiya T, Danku J, Naseer S, Geldner N,Guerinot M L, Salt D E. 2013. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root.Proceedings of the National Academy of Sciencesof the United States of America,110, 14498–14503.
Kaufmann M J, Gerdemann J. 1958. Root and stem rot of soybean caused byPhytophthora sojaen.sp.Phytopathology,48, 201–208.
Koenning S R, Wrather J A. 2010. Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009.Plant Health Progress,10, doi: 10.1094/PHP-2010-1122-01-RS
Kosma D K, Murmu J, Razeq F M, Santos P, Bourgault R,Molina I, Rowland O. 2014. AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types.The Plant Journal,80, 216–229.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plantcis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences.Nucleic Acids Research,30, 325–327.
Li Q, Chen J, Xiao Y, Di P, Zhang L, Chen W. 2014. The dirigent multigene family in Isatis indigotica: Gene discovery and differential transcript abundance.BMC Genomics,15, 388.
Lin F, Zhao M, Ping J, Johnson A, Zhang B, Abney T S,Hughes T J, Ma J. 2013. Molecular mapping of two genes conferring resistance toPhytophthora sojaein a soybean landrace PI 567139B.Theoretical and Applied Genetics,126, 2177–2185.
Link A J, Eng J, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R. 1999. Direct analysis of protein complexes using mass spectrometry.Nature Biotechnology,17, 676–682.
Ma Q H, Liu Y C. 2015. TaDIR13, a dirigent protein from wheat,promotes lignan biosynthesis and enhances pathogen resistance.Plant Molecular Biology Reporter,33, 143–152.
Ping J, Fitzgerald J C, Zhang C, Lin F, Bai Y, Wang D, Aggarwal R, Rehman M, Crasta O, Ma J. 2016. Identification and molecular mapping ofRps11, a novel gene conferring resistance toPhytophthora sojaein soybean.Theoretical and Applied Genetics,129, 445–451.
Ralph S, Park J Y, Bohlmann J, Mansfield S D. 2006. Dirigent proteins in conifer defense: Gene discovery, phylogeny,and differential wound-and insect-induced expression of a family of DIR and DIR-like genes in spruce (Piceaspp.).Plant Molecular Biology,60, 21.
Ralph S G, Jancsik S, Bohlmann J. 2007. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny,and constitutive and stress-induced gene expression in spruce (Piceaspp.).Phytochemistry,68, 1975–1991.
Rocco M, Arena S, Renzone G, Scippa G S, Lomaglio T,Verrillo F, Scaloni A, Marra M. 2013. Proteomic analysis of temperature stress-responsive proteins inArabidopsis thalianarosette leaves.Molecular BioSystems,9, 1257–1267.
Shen C Y, Su Y C. 1991. Discovery and preliminary studies ofPhytophthora megaspermaon soybean in China.Acta Phytopathologica Sinica,4, 017. (in Chinese)
Stephens D J, Lin-Marq N, Pagano A, Pepperkok R, Paccaud J P. 2000. COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity to ER exit sites.Journal of Cell Science,113, 2177–2185.
Sugimoto T, Yoshida S, Kaga A, Hajika M, Watanabe K, Aino M, Tatsuda K, Yamamoto R, Matoh T, Walker D. 2011.Genetic analysis and identification of DNA markers linked to a novelPhytophthora sojaeresistance gene in the Japanese soybean cultivar Waseshiroge.Euphytica,182, 133.
Sun S, Wu X L, Zhao J M, Wang Y C, Tang Q H, Yu D Y,Gai J Y, Xing H. 2011. Characterization and mapping ofRpsYu25, a novel resistance gene toPhytophthora sojae.Plant Breed,130, 139–143.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Molecular Biology and Evolution,28, 2731–2739.
Thazar-Poulot N, Miquel M, Fobis-Loisy I, Gaude T. 2015.Peroxisome extensions deliver theArabidopsisSDP1 lipase to oil bodies.Proceedings of the National Academy of Sciences of the United States of America,112, 4158–4163.
Tian M, Zhao L, Li S, Huang J, Sui Z, Wen J, Li Y. 2016.Pathotypes and metalaxyl sensitivity ofPhytophthora sojaeand their distribution in Heilongjiang, China 2011–2015.Journal of General Plant Pathology,82, 132–141.
Vanrobaeys F, Van Coster R, Dhondt G, Devreese B, Van Beeumen J. 2005. Profiling of myelin proteins by 2D-Gel electrophoresis and multidimensional liquid chromatography coupled to MALDI TOF?TOF mass spectrometry.Journal of Proteome Research,4, 2283–2293.
Wang Q, Han C, Ferreira A O, Yu X, Ye W, Tripathy S, Kale S D,Gu B, Sheng Y, Sui Y. 2011. Transcriptional programming and functional interactions within thePhytophthora sojaeRXLR effector repertoire.The Plant Cell,23, 2064–2086.
Wang X, Fan C, Zhang X, Zhu J, Fu Y F. 2013. BioVector, a flexible system for gene specific-expression in plants.BMC Plant Biology,13, 198.
Weidenbach D, Esch L, M?ller C, Hensel G, Kumlehn J, H?fle C, Hückelhoven R, Schaffrath U. 2016. Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modular poaceae-specific proteins.Molecular Plant,9, 514–527.
Weng C, Yu K, Anderson T, Poysa V. 2001. Mapping genes conferring resistance toPhytophthoraroot rot of soybean,Rps1aandRps7.Journal of Heredity,92, 442–446.
Wu R, Wang L, Wang Z, Shang H, Liu X, Zhu Y, Qi D, Deng X. 2009. Cloning and expression analysis of a dirigent protein gene from the resurrection plantBoea hygrometrica.Progress in Natural Science,19, 347–352.
Wu X I, Zhang B Q, Shi S, Zhao J M, Feng Y, Na G, Gai J Y,Han X. 2011. Identification, genetic analysis and mapping of resistance toPhytophthora sojaeof Pm28 in soybean.Agricultural Sciences in China,10, 1506–1511.
Xu G, Ma H, Nei M, Kong H. 2009. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification.Proceedings of the National Academy of Sciences of the United States of America,106, 835–840.
Xu W, Yu Y, Ding J, Hua Z, Wang Y. 2010. Characterization of a novel stilbene synthase promoter involved in pathogenand stress-inducible expression from Chinese wildVitis pseudoreticulata.Planta,231, 475.
Yang B, Wu J, Gao F, Wang J, Su G. 2014. Polyamine-induced nitric oxide generation and its potential requirement for peroxide in suspension cells of soybean cotyledon node callus.Plant Physiology and Biochemistry,79, 41–47.
Yao H Y, Wang X M, Wu X F, Xiao Y N, Zhu Z D. 2010.Molecular mapping ofPhytophthoraresistance gene in soybean cultivar Zaoshu 18.Journal of Plant Genetic Resources,11, 213–217.
Zhang J, Xia C, Wang X, Duan C, Sun S, Wu X, Zhu Z. 2013.Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar.Theoretical and Applied Genetics,126, 1555–1561.
Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L,Zhao Y, Ma Y, Fang C, Shen Y, Liu T, Li C, Li Q, Wu M,Wang M, Wu Y, Dong Y, Wan W,et al. 2015. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean.Nature Biotechnology,33, 408–414.
Zipfel C. 2014. Plant pattern-recognition receptors.Trends in Immunology,35, 345–351.
 Journal of Integrative Agriculture2018年6期
Journal of Integrative Agriculture2018年6期
- Journal of Integrative Agriculture的其它文章
- Improve access to the EU market by identifying French consumer preference for fresh fruit from China
- Management and prevention of mastitis: A multifactorial approach with a focus on milking, bedding and data-management
- Elimination of ceftiofur hydrochloride residue in postpartum cows’milk after intramammary infusing at dry-off
- Evaluation of a new qPCR test to identify the organisms causing high total bacterial count in bulk tank milk
- Prevalence and characteristics of extended spectrum β-lactamaseproducing Escherichia coli from bovine mastitis cases in China
- Evolutionary analysis of plant jacalin-related lectins (JRLs) family and expression of rice JRLs in response to Magnaporthe oryzae
