Evolutionary analysis of plant jacalin-related lectins (JRLs) family and expression of rice JRLs in response to Magnaporthe oryzae
State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops/Key Laboratory of Biopesticide and Chemistry Biology, Ministry of Education/College of Life Science, Fujian Agriculture and Forestry University, Fuzhou 350002, P.R.China
1. Introduction
Lectins, a group of proteins that have at least one noncatalytic carbohydrate binding site, bind reversibly to specific mono- or oligo-saccharides (Peumans and van Damme 1995; Vandenborreet al. 2011). Lectins have diverse functional roles in plants and animals (Peumans and van Damme 1995). Plant lectins can be classified into 12 sub-families (Peumanset al. 2001). Among them, jacalin proteins were first isolated from seeds of jack fruit (Moreira and Ainouz 1981). So far, at least 25 jacalin-related lectins(JRLs) have been identified in plants (Table 1). Among these, nine are chimeric JRLs that contained the exogenous domain dirigent which has important roles in plant secondary metabolism (Hosmaniet al. 2013). The remaining 16 are merolectins or hololectins according to the classification scheme for lectins (Peumans and van Damme 1995).
During the evolution, plants have to overcome various environmental challenges at different phases of their life cycles (Yamajiet al. 2012; Sandalioet al. 2015; Nejatet al.2017; Sahayet al. 2017). Duplication and rearrangement of adaptive genome modules play an important role for this process. Such events have increased the number ofNBS-LRR(nucleotide- binding site plus leucine-rich repeat)genes involved in resistance (Richlyet al. 2002). The plantlectin superfamily shows species-specific gene expansion caused by tandem/segmental duplications. InArabidopsis,soybean, and rice, the tandem and segmental duplications lead to lectin gene expression divergences under biotic and abiotic stresses (Jianget al. 2010).In wheat, JRLs experienced a substantial diversification after the divergence of wheat from other cereal species (Songet al. 2014).
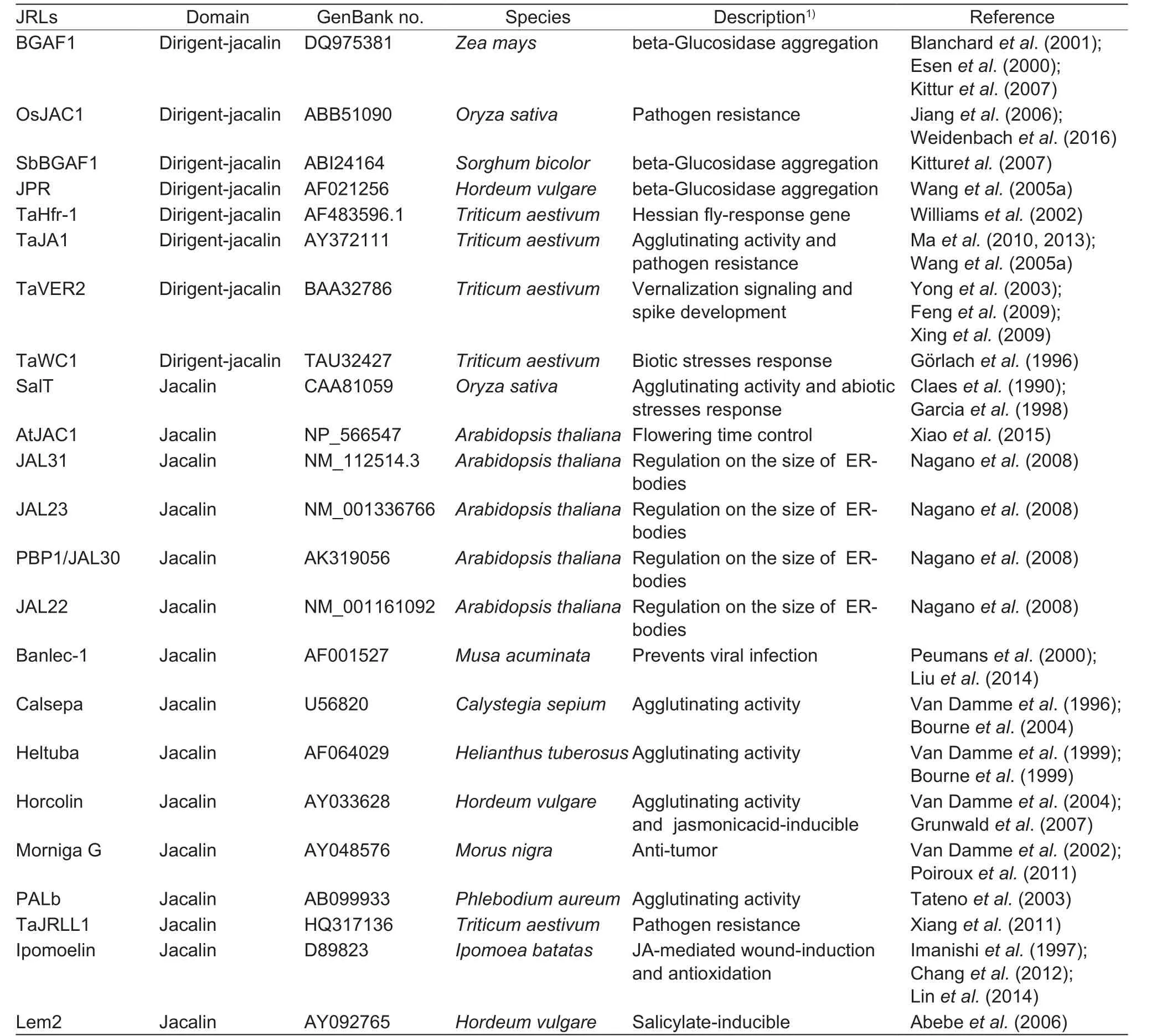
Table 1 Summary on already known plant jacalin-related lectins (JRLs)
JRLs are associated with plant responses to environmental stresses and pathogen attacks. At the gene level, quite a number of wheatJRLsgenes are stress-inducible and tissue-specific (Songet al. 2014). The wheat JRL proteinTaWC1is an inducible gene that is up-regulated as part of the wheat acquired resistance response (G?rlachet al.1996). Mannose-binding jacalinTaHfr-1is transcriptionally induced in wheat by Hessian fly larval infection (Williamset al. 2002). Another mannose-specific jacalin-related lectin-like gene (TaJRLL1) is induced by pathogen infection,as well as salicylic acid (SA) and jasmonic acid (JA)inducing treatments. Expression ofTaJRLL1inArabidopsisenhances resistance against fungal diseases (Xianget al.2011). Transient expression of the wheat JRL geneTaJA1in barley also confers resistance against the powdery mildew fungus (Weidenbachet al. 2016). In rice, SalT protein was first isolated from salt-treated rice tissue of theindicavariety Taicheng Native 1 (Claeset al. 1990). TheSalTgene shows organ-specific expression and is induced by drought and hormone treatments.Another jacalin-related lectin protein OsJAC1 accumulates at fungal penetration sites and confers resistance against various rice pathogens (Jianget al.2006; Weidenbachet al. 2016). Jacalin proteins not only recognize carbohydrates for defense in plant, but also have roles during developmental processes. For example, theArabidopsisjacalin-like geneAtJAC1is a positive regulator in controlling timing of plant flowering (Xiaoet al. 2015).Two pairs of heterodimer-stype jacalins (JAL31-JAL23 and JAL30-JAL22) have opposite effects on regulating the size of the PYK10 complex which serves as a major part of endoplasmic reticulum (ER) bodies and may be involved inArabidopsisdefense systems (Naganoet al. 2008).
Although 28 JRLs have been predicted in the rice genome and some of the genes are responsive to various stresses(Jianget al. 2010), details on gene composition, architecture,regulation, and function of rice JRLs are still unclear. In this study, we profiled and compared evolution and diversification of JRLs in 30 plant species. We found that rice contains sevenPoaceae-specific JRLs by fusion with a dirigent domain or a NB_ARC domain. It is possible that the chimeric jacalins play specific roles in rice defense response against pathogens.The subsequent gene expression assays show that four of thePoaceae-specificOsJRLs genes as well as 19 of the mero-/holo-jacalins have positive responses by being induced afterMagnaporthe oryzaeinoculation, indicating that these JRLs can indeed be an important part of the rice immune system directed againstM.oryzae.
2. Materials and methods
2.1. ldentification of jacalin domain proteins in plant genomes
Genome wide protein sequences ofAnanas comosus,Aquilegia coerulea,Brachypodium distachyon,Brassica oleracea,Brassica rapa,Carica papaya,Medicago truncatula,Mimulus guttatus,Physcomitrella patens,Populus trichocarpe, andSelaginella moellendorffiiwere downloaded from JGI Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). ForBrassica napus,Glycine max,Sorghum bicolor,Triticum aestivum,Vitis vinifera,andZea mays, the genome wide protein sequences were downloaded from Ensembl database (http://plants.ensembl.org/index.htm). Genome wide amino acid sequences ofGossypium hirsutum,Gossypium arboreum,andGossypium raimondiiwere downloaded from Cottongen (https://www.cottongen.org/). ForNicotiana benthamiana,Solanum tuberosum,andSolanum lycopersicum, genome wide protein sequences were downloaded from Sol Genomics Network Search (http://solgenomics.net/).Arabidopsis lyratagenome was downloaded from JGI Genome Portal (https://genome.jgi.doe.gov/Araly1/Araly1.home.html). Genome wide amino acid sequences ofArabidopsis thalianawere downloaded from TAIR (http://www.arabidopsis.org/). Genome wide amino acid sequences ofElaeis guineensiswere downloaded from Eukaryotic Genome Annotation at NCBI(http://www.ncbi.nlm.nih.gov/genome/annotation_euk).Genome wide amino acid sequences ofHordeum vulgarewas downloaded from MIPS Barley Genome Database(http://pgsb.helmholtzmuenchen.de/plant/ plantsdb.jspnfa). Genome ofLotus japonicuswas downloaded from PlantGDB (https://plantgdb.org/LjGDB). Genome wide amino acid sequences ofMusa nanawas downloaded fromBanana Genome Hub (http://banana-genomehub.southgreen.fr/). All annotated amino acid sequences ofOryza sativawere downloaded from RGAP MSU rice genome project database (http://rice.plantbiology.msu.edu/index.shtml).
For those protein sequences, jacalin domain (PF01419.14)was then identified using the Hmmerscan Program in HMMER v3.1b2 with default settings (http://hmmer.org/).The obtained sequences were further manually compared with BlastP (E value 1e-10) citation (Altschulet al. 1997;Mistryet al. 2013).
2.2. Phylogenetic tree construction
Multiple alignments of the full length sequences of JRLs were made using fast multiple sequence alignment software MAFTT (Multiple Alignment using Fast Fourier Transform)(Katoh and Standley 2013) with default parameters. For the large-scale alignment, the fast maximum-likelihood phylogenetic tree (Figs. 1–4) were generated using FastTree by JTT+CAT model (Priceet al. 2009; Liuet al. 2011). In the phylogenetic tree, supporting values above 50 were kept and regarded reliable. Tree in Fig. 1-A was draw by Figtree and other trees in this paper were drawn by online itol tree(http://itol.embl.de/#).
2.3. Orthologous JRLs protein group
Amino acids sequences from 30 plant species were used for an Orthofinder search (Emms and Kelly 2015) using default settings. The orthologous genes from each protein family were listed by group(s) (Appendices A–C).
2.4. Plant growth and inoculation assay
Rice plants were grown in a growth chamber at 28°C with 16 h light and 8 h darkness.M.oryzaeconidia were produced by growing cultures on rice bran medium (4%rice bran, 2% agar, pH 6.0) for 5 days in the dark followed by exposure to 12 h light/dark cycles for further 2 days at 25°C (Chenet al. 2008). For the plant inoculation assays,conidia were harvested from the rice bran plate cultures withM.oryzaeisolates and suspended to a concentration of 1×105spores mL–1in 0.2% Tween 20 in distilled water.Plants at the four-leaf-stage were then spray-inoculated with conidia suspension (Chenet al. 2008). Rice leaf samples were harvested at 0, 24, and 48 hours post inoculation (hpi).
2.5. Expression profile and quantitative RT-PCR analysis
For RNA-seq, publicly available RNA-seq data forM.oryzaeand rice interaction were obtained from NCBI GEO database(www.ncbi.nlm.nih.gov/geo/). Sequencing reads were mapped to rice reference sequence with Tophat, Cufflinks,and Cuffmerge to extract all possible exons (Trapnellet al.2012). Expression abundance were calculated by RSEM with default parameters (Liet al. 2011). The RPKM values were log transformed, and genes were clustered according to their expression patterns using the R package Pheatmap.

Fig. 1 Phylogenetic relationship of jacalin-related lectins (JRLs) of rice and 29 other plant species. A, the phylogenetic tree was divided into four clades, each represented by different colors of the surrounding dots. Blue, yellow, purple, and green represent clades 1, 2, 3, and 4, respectively. B, the 15 leaf colors are used to distinguish the 15 families that shown in the columns below the tree.
For real-time PCR, the total RNAs were extracted using RNAiso Plus (TaKaRa, China) and quantified by Nanodrop(Thermo, USA). The first chains of cDNA were synthesized using ReverTraAce qPCR RT Master Mix with gDNA Remover Kit (TOYOBO, Japan) according to manufacturer’s instructions. Quantitative RT-PCR amplifications were performed in 10 μL reactions containing 5 μL of 2×SYBR PremixExTaq(TliRNaseH plus, TaKaRa), 10 ng of cDNA template and 0.75 μL of each primer (10 μmol L–1). The reactions were then subjected to Eppendorf Mastercycler Realplex Real-Time PCR System (Eppendorf, Germany).The reaction settings were 95°C for 5 min; 40 cycles of 95°C for 20 s, 56°C for 15 s. The cycle threshold value of each gene was normalized to the internal reference geneOsActin.The relative expression of each gene was calculated by the 2–ΔΔCtmethod (Livak and Schmittgen 2001; Schmittgen and Livak 2008).
3. Results
3.1. ldentification and phylogeny of JRLs in 30 selected plant species
To analyze the evolutionary relationships of JRLs, combined BlastP and Pfam Search Program which were used to identify JRLs in 30 different plants across 15 monocots-/dicots-families. A total of 651 JRLs have been identified in 30 plant species (Fig. 1-B). It’s noticeable the total number of JLRs varied in different plant species and this variation is not related with genome size. The largest two families includeBrassicaceaewith 324 JRLs andPoaceaewith 157 JRLs.The number of JRLs per species inBrassicaceaeranges from 46 to 123, while inPoaceaefrom 20 to 41. The lowest number is found inC. papayathat only contains two JRLs.
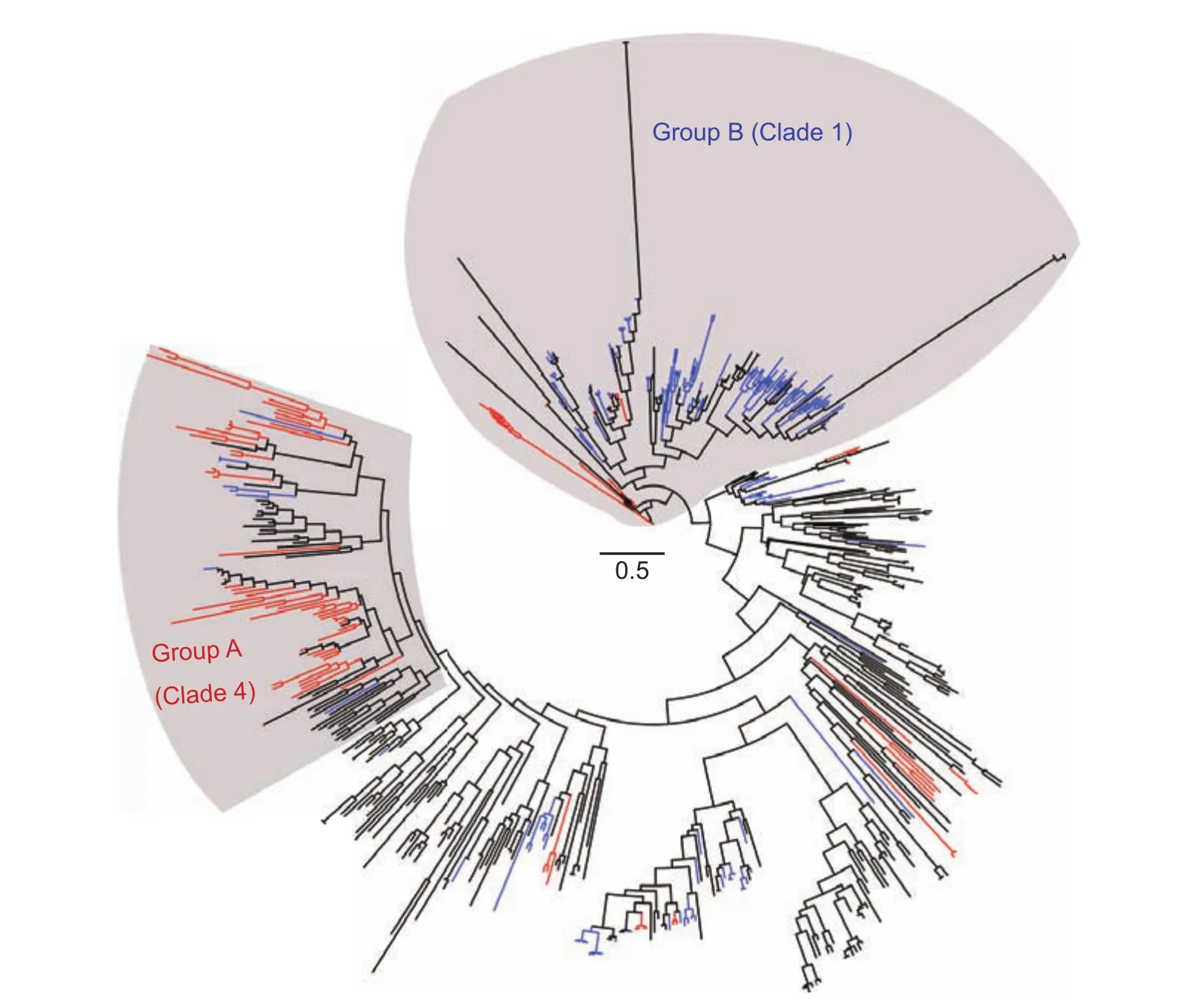
Fig. 2 Branch-length view of phylogenic tree of jacalin-related lectins (JRLs) from rice and 29 other plant species. Chimeric jacalins,holojacalins, and merojacalins are shown with red, blue, and black branches, respectively.
Classic JRLs have only one jacalin domain, while JRLs proteins identified in this study contain three sub-types with the following domain-combinations: merojacalin, holojacalin,and chimeric jacalin, according to the classification of lectin (Peumans and Damme 1995). Merojacalin contains only one jacalin domain. Holojacalin contains at least two identical jacalin domains without fusion with other domains. Chimeric jacalin have one or more jacalin domains that combine(s) with other types of domains such as dirigent, Pkinase, NB_ARC or Kelch_1/2/3/6 found in this study (Appendix D). The lower plantP. patens(a moss) has only seven JRLs, while five out of the seven JRLs are chimeric jacalin with Pkinase_Tyr domain which is otherwise mainly found inPoaceaespecies. In the fern plantS. moellendorffi, seven out of eight JRLs are merojacalins while the last one is a holojacalin. In higher plants,Solanaceaespecies have merojacalin as the major sub-type (21 out of 39), and then holojacalin (12 out of 39).Besides merojacalin and holojacalin,N. benthamianahas three chimeric jacalins with NB_ARC domain which is absent in tomato or potato.Leguminosae,Malvaceae,A. coerulea,M. guttatus,V. vinifera,C. papaya, andP. trichocarpacontain less number of jacalins (from two to 10) and have different distributions of merojacalin and holojacalin. For example, inLeguminosae, soybean also has three sub-type jacalins, two for each type of jacalin proteins.M. truncatulacontains only one merojacalin and one holojacalin.L. japonicus has six merojacalins, one holojacalin, and two chimeric jacalins with DIOX_N and OG-FeII_Oxy domains.The last appears conserved at the N-terminal region by containing 2-oxoglutarate/Fe(II)-dependent dioxygenase that catalyzes O-demethylation (Hagel and Facchini 2010).In five selectedBrassicaceaespecies,more than half (about 55%) of JRLs are holojacalins. Merojacalins are the second most abundant with 28%. The remaining JRLs are chimeric jacalins that mainly contain F-box associated (FBA) domain and Kelch-repeat domain with 3 and 11%, respectively.For monocot species, the major subtype is merojacalin.A. comosus,M. nana, andE. guineensishave two subtype jacalins: merojacalin and holojacalin with proportions 11:4,20:1, and 10:4, respectively. InPoaceae, JRLs are present as two main subtypes: merojacalins (50%) and chimeric jacalins (40%), while holojacalins only constitutes 10%.
A phylogenetic maximum-likelihood tree was generated based on a multiple alignments of full-length amino acid sequences of JRLs by FastTree to investigate evolutionary relationships (Fig. 1-A). The phylogeny of 651 JRLs resolves the four expected groups. The majority ofBrassicaceaeJRLs are included in clade 1 (Fig. 1-A) along with a fewPoaceaeJRLs. We further made the alignments of JRLs in this clade and found thatPoaceaeJRLs shared high value of 20–83% identity with certainBrassicaceaeunique JRLs (Appendix E), suggesting that horizontal gene transfer and homoplasy may exist betweenBrassicaceaeandPoaceae. In clade 2 (Fig. 1-A),BrassicaceaeJRLs are nested withP. trichocarpa,M. guttatus, andA. coerulea.Clade 3 (Fig. 1-A) contains JRLs from all 30 examined species, including moss, fern, dicots, and monocots, and seems evolutionally conserved, indicating that JRLs in this clade could have been present before the divergence of monocots and dicots. JRLs in clade 4 are only present within asPoaceaespecies (Fig. 1-A).
To further describe the distribution of different types of JRLs in 30 selected genomes, we made a more detailed view of the distribution of merojacalins, holojacalins, and chimeric jacalins (Fig. 2) based on the data presented in Fig. 1.PoaceaeJRLs in clade 4 (Fig. 1-A) are highly prone to have chimeric jacalins (55%) compared to merojacalins(37%) and holojacalins (8%) (Fig. 2, Group A). The integrated domains of these jacalin proteins are listed in Table 2 and mainly found to be Pkinase, dirigent, and NB_ARC domains. In contrast, JRLs in clade 1 (Fig. 1-A)mainly contain holojacalins (60%), followed by merojacalins(20%) and chimeric jacalins (20%) (Fig. 2, Group B). From dicots to monocots, chimeric jacalins in clade 4 show more evolutionary differentiations than the other clades, which could be a special adaption inPoaceaespecies. These results suggest the number and domain combination of JRLs in different plant species are highly variable and JRLs inPoaceaespecies have experienced different diversification compared withBrassicaceaespecies.
3.2. Evolution and divergence of JRLs in Poaceae
To further characterize JRLs inPoaceae, we investigated the phylogeny of jacalin-related lectins in grass species. A maximum likelihood phylogenic tree is constructed by using the full-length amino acid sequences. Six groups of JLRs are categorized by containing representative domains from six grass species, includingO. sativa,Z. mays,H. vulgare,T. aestivum,S. bicolor, andB. distachyon(Fig. 3).
Clade 6 (Fig. 3) contains 6 wheat and barley JRLswith four types of domain integrations: merojacalins (20%),holojacalins (50%) and chimeric jacalins (30%) among which the integrated domains are fused to single-jacalin domains(10%) and multiple-jacalin domains (20%) (Table 3). Clade 1(Fig. 3) is the largest group by consisting of 60 JRL proteins across all six species: six from maize, eight from sorghum, 10 from wheat, 11 fromB.distachyon, 11 from barley, and 14 from rice. Of the JRLs located in clade 1(Fig. 3), 80% contain a single jacalin domain, compared with 13% containing repeated jacalin domains (Table 3).Only two JRLs (Os11g06570 and Os04g12390) contain other domains, such as Peptidase_C43, Peptidase_C48,DUF4216, DUF4218, or Transposase domains. Similarly,most of JRLs in clade 3 (Fig. 3) are found coding for only one jacalin domain (94%) except for wheat JRL coding for one jacalin containing Pkinase-jacalin-jacalin domains(Traes_4AL_015A5C163.3) that is found as a chimeric jacalin (Fig. 3, leaves labeled with dash lines). The other three groups (Fig. 3) (clades 2, 4, and 5) all contain chimeric jacalins. Most of these proteins encode for other types of domains, such as dirigent, Pkinase, and NB_ARC.
Clade 5 (Fig. 3) contains 10 chimeric jacalins and one merojacalin that code for only one single jacalin domain across four cereal species but is not present inB.distachyonand barley: one from sorghum, two from wheat, three from maize, and five from rice. Chimeric jacalins in this clade are present as jacalin-NB_ARC and multiple jacalins-NB_ARC,which comprise 45 and 45% of all jacalins, respectively(Table 3). This clade forms two paired monophyletic subgroups present as paralogs: Os11g39480–Os11g39530 and GRMZM2G355098_P01–GRMZM2G368663_P01. The maize pair with a high bootstrap (100%) assembles jacalin and NB_ARC in a similar way, indicating a duplication event occurred before the maize-rice divergence. The rice pair did not share much common in domain arrangements.Os11g39530 contains two jacalin domains and one NB_ARC domain while Os11g39480 has only one jacalin domain,which may due to an occasional loss during duplications.Clade 4 (Fig. 3) is composed of 10 JRLs proteins, one fromB.distachyon, one from sorghum, two from barley, three from rice, and three from wheat. Eight of the chimeric jacalins harbor a kinase domain across all the grass species.A Pkinase domain is fused to single (50%) or multiple (30%represented) jacalin domains among all leaves in this clade.The other JRLs, Br_4g13990 and Traes_5BS_60FAB49FB.1,encode for three and one jacalin domains, respectively,without any exogenous domains. Two monophyletic pairs Traes_2AL_F5E021100.1–Traes_2DL_59C23B50A.2 and Os11g32210.1–Os11g32170.1 are formed in this clade.Traes_2AL_F5E021100.1–Traes_2DL_59C23B50A.2 share high amino acid sequence identity which may be a result of segmental duplication events which also occurred in the Os11g32210.1–Os11g32170.1 pair.
Clade 2 (Fig. 3) is comprised of 48 JRLs proteins from six grass species: three from barley, five from rice, eight from sorghum, nine from maize, 11 fromB.distachyon,and 12 from wheat. Maize JRLs cluster in two sub-clans(M1 and M2). The M1 proteins are closely related to the sorghum JRL Sb09g001880 and the M2 proteins are related to Sb02g007740, Sb02g007735, Sb02g007750,and Sb02g007760 (Fig. 3). Those JRLs are more related to each other than to JRLs from the same species from other clades, suggesting that maize and sorghum have a common ancestor and the duplication already existed prior the divergence of maize-sorghum. A sub-branch containing rice JRLs (including Os12g09720.1, Os12g09700.1,Os12g14440.1, and Os12g1270.1) is located closely to maize-sorghum. Duplication of the rice JRLs in this subbranch may have occurred before the ancestor of maizesorghum separated from rice. In contrast, the wheat subbranch W1 JRLs probably have duplicated more recently than the divergence of wheat from other grass species(Fig. 3).
Domain analysis showed that 39 out of 48 JRLs encodes for dirigent-jacalin chimeric proteins. In all these JRLs,the dirigent domain is fused with only a single jacalin domain. Using protein sequence clustering analysis, we found a total 53 of JRLs are only found in the six grass species (Fig. 3). A total of 40 of the grass-specific JRLs that contain dirigent domain are found in clade 2 (Fig. 3).The remaining eight JRLs are found in clade 4 and contain a Pkinase domain (Fig. 3). In a group comparison, there is a particularly high number of grass-specific JRLs found in Group A (Fig. 2) that contains dirigent domain jacalins(dash leaves with stars in clade 2 or 4 of Fig. 4). Rice JRLs (Os02g19890.1, Os11g32170.1, Os11g32210.1,Os12g09720.1, Os12g09700.1, Os12g14440.1, and Os12g12720.1) are only found in grass species. Together,our results indicate that a rapid domain recombination of JLRs has occurred in Poaceae, and a unique group has been identified in rice genome.
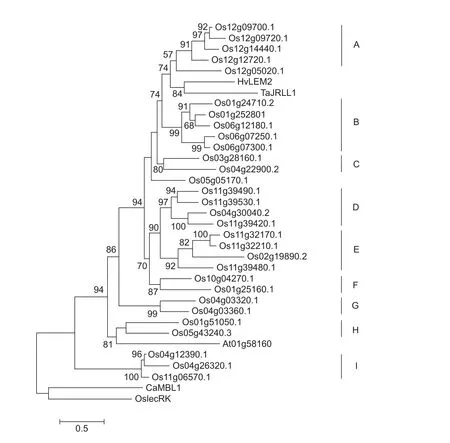
Fig. 4 Phylogeny of rice jacalin-related lectins (JRLs). The phylogenetic tree was constructed based on multiple alignments of full-length amino acid sequences using Multiple Alignment using Fast Fourier Transform (MAFFT). The resulting alignment was then used to generate a maximum-likelihood tree by FastTree. The scale bar shows 0.5 amino acid substitutions per site. CaMBL1 and OslecRK are set as out-group. Rice JRLs are divided into nine subgroups, from A to I.
3.3. Jacalin-related lectins in rice
Comparison of JRLs inPoaceaeindicates the rice genome contain a unique group of dirigent domain jacalins. We next focus on rice and analyze phylogeny of rice JRLs. In this species, two more jacalins are found than those presented in the report by Jianget al. (2010). All JRLs in rice (OsJRLs)contain full open reading frames and are distributed on nine of 12 chromosomes, with none found on chromosomes 7 to 9 (Table 4). The deduced protein products of the 30 OsJRLs range from 12.2 to 73.5 kDa with pI value from 4.8 to 9.4. All OsJRLs contain typical jacalin domain structures and some jacalin domains are fused to other types of domains. Most of the rice JRLs (16 out of 30) have only one jacalin domain as merojacalins. Thirteen out of 30 OsJRLs are chimeric proteins that have recruited NB_ARC, Pkinase, dirigent or Peptidase domain to the jacalin domain(s) and cluster on chromosome 11 or 12 (Table 4). Only one OsJRLs is a holojacalin and it contains three repeated jacalin domains.
In order to evaluate the evolutionary relationship among the 30 OsJRLs, we performed a phylogenetic analysis based on full amino acid sequences and compared withbarley HvLEM2, wheat TaJRLL1,ArabidopsisAt01g58160,and pepper CaMBL1 (Fig. 4). The last two are used as out-group. Nine clades (A–I) (Fig. 4) are identified as monophyletic subgroups with high bootstrap (≥80%),suggesting that OsJRLs in the same clade may share the same evolutionary origin. The additional two JRLs(Os12g05020 and Os05g05170) form single branches.Different types of jacalins distributes into subfamilies.In subfamilies B, C, F, G, and H, OsJRLs are present as merojacalins. Subfamilies A, D, and E are enriched in chimeric jacalins with dirigent, Pkinase or NB_ARC domain, respectively. In addition, seven sister pairs paralogous with rice JRLs are found with strong bootstrap(≥92%), Os12g09700.1–Os12g09720, Os06g07250.1–Os06g7300.1, Os11g39490.1–Os11g39530.1,Os4g30040.2–Os11g39420.1, Os11g32170.1–Os11g32210.1, Os04g03320.1–Os04g03360.1, and Os04g12390.1–Os04g26320.1, indicating that a clear paralogous pattern of JRLs divergence caused by duplication occurs in rice.
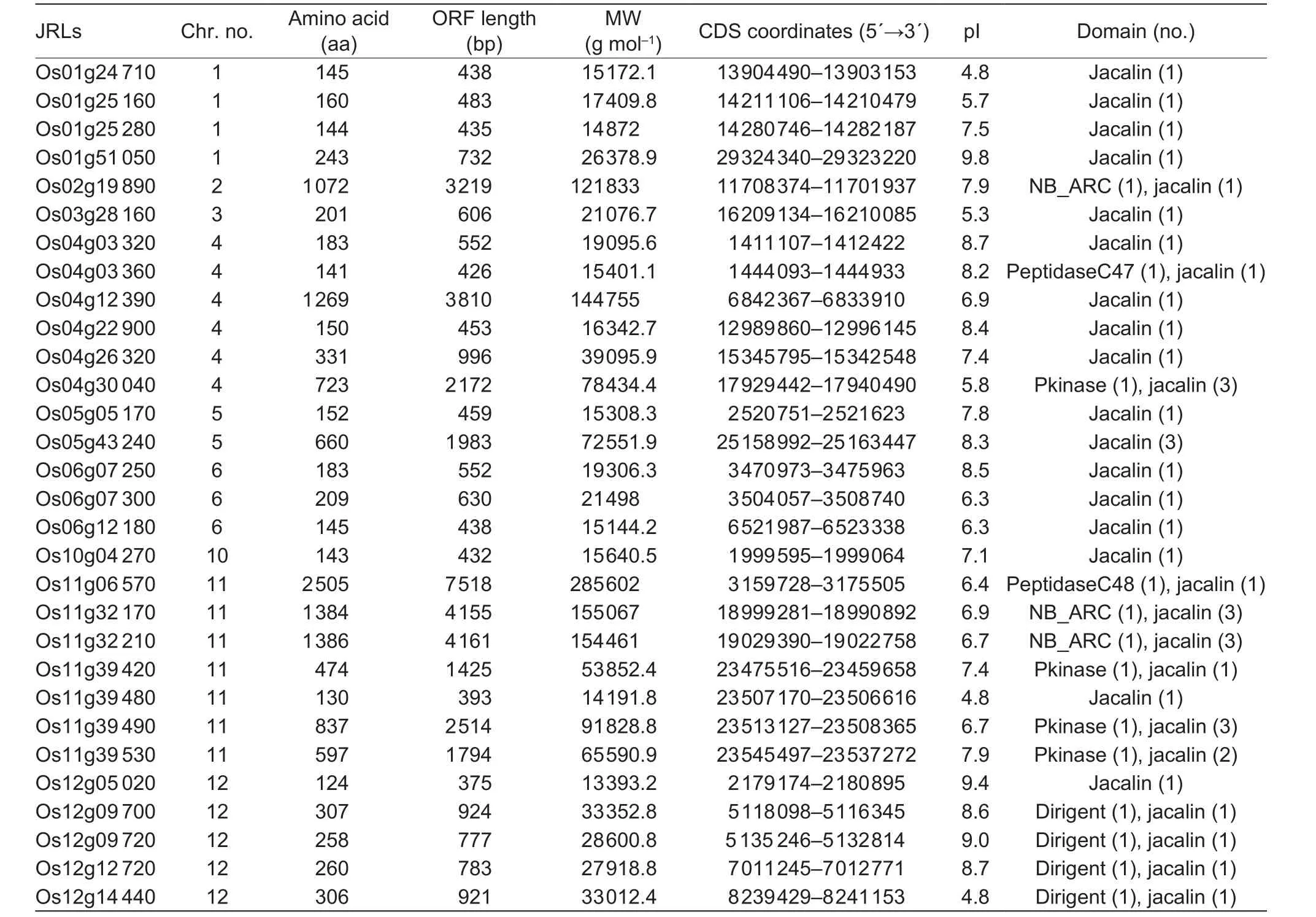
Table 2 Characteristics of jacalin-related lectins (JRLs) from rice genome1)

Table 3 Distribution of grass chimeric jacalins in group A of Fig. 3

Table 4 Summary of jacalin domains found in each clade (Fig. 4) of grass phylogeny
3.4. Expression patterns of rice JRLs genes in response to rice blast disease
Gene expression patterns offer important clues of gene functions. To investigate possible involvement of JRLs in the response toM.oryzae, we examined the expression ofOsJRLsgenes in rice duringM.oryzaeinfection (at time 0, 24 and 48 hpi) using RNA sequencing. TwoM.oryzaeisolates (Guy11 and FJ81278) are used to inoculate the susceptible rice cultivar Nipponbare in this assay. Most of theOsJRLsshow detectable expression in the assay except seven genes (Os01g25160, Os04g03320, Os04g03360,Os11g06570, Os11g32170, Os11g39480, and Os12g12720)(Fig. 5). The inducible genes include 12 merojacalins, one holojacalin, and 10 chimeric jacalins. Of the 10 chimeric jacalins, three arePoaceaespecific dirigent-jacalins. Of the remaining, four chimeric jacalins contain NB_ARC and three contain Pkinase domains. The expression patterns of the three types of jacalins in rice are different. The 19OsJRLs are divided into six groups depending on their expression patterns. Compared to the control treatment,Guy11 and FJ81278 strains up-regulate most of theOsJRLs in group 1 (Os5g43240 and Os12g09700) and group 2(Os10g04270, Os11g39490, and Os01g25280) at 48 hpi.Group 3 (Os04g12390 and Os03g28160) shows marked down-regulation at 24 hpi for both Guy11 and FJ81278 inoculations but a slower return to control levels at 48 hpi for FJ81278 than that for Guy11. Group 5 (Os01g51050,Os12g05020, Os04g26320, and Os04g22900) are downregulated byM.oryzaeinfections but with slower response in the FJ81278 treatment.OsJRLsin Group 4 (Os06g07300 and Os06g12180) are strongly up-regulated by FJ81278but not by Guy11. Group 6 JRLs are up-regulated at 24 hpi and then returns down to control levels at 48 hpi with a stronger up-regulation for Guy11 inoculated rice.
To verify these results, eightOsJRLs genes were then examined by real-time PCR for their expression in Nipponbare rice after inoculation withM.oryzaeGuy11.These eight gene were responsive toM.oryzaechallenge mainly at 24 or 48 hpi. The transcriptional profiles of Os01g24710, Os06g07250, and Os11g32210 are all elevated after 12 hpi and reached the maximum at 24 hpi.Os12g14440 (also known as OsJAC1) has a much earlier response and reach the peak at 12 hpi, which is consistent with the finding by Jiang (2006) and Weidenbach (2016).Os01g25280, Os05g43240, Os10g04270, and Os12g09720 have similar expression pattern by being up-regulated at 48 hpi.
4. Discussion
4.1. Phylogeny of plant JRLs
Jacalin-related lectin proteins are found in many plants,some of which have been characterized and take roles in enhancing tolerance against environmental stresses and pathogens attacks (Xiaoet al. 2005; Jianget al. 2006; Xianget al. 2011; Maet al. 2013; Weidenbachet al. 2016). In this study, we increase the number of studied plants since more genomes are now available and analyzed the distribution,phylogenic relationship, and domain composition of JRLs in plants across 15 monocots-/dicots-families.
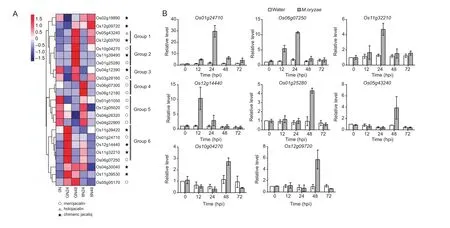
Fig. 5 Expression profiles of rice jacalin-related lectins (OsJRLs) following rice blast inoculation. A, heat map of 23 OsJRLs genes regulated by Magnaporthe oryzae inoculations. Color represents relative expression level for RNA sequencing. Scale ranges from blue (low) to red (high). Up-regulation (dark red) and down-regulation (dark blue) are described by log2fold changes relative to the mean expression of the selected genes in three stages: uninoculated rice (0N), inoculated with Guy 11 at 24 hours post inoculation (hpi) (GN24), inoculated with Guy11 at 48 hpi (GN48), inoculated with FJ81278 at 24 hpi (8N24), and inoculated with FJ81278 at 48 hpi (8N48). B, qRT-PCR expression of eight selected OsJRLs genes in susceptible rice in exposure to Guy 11.Error bars represent the standard deviation from three replications.
JRLs are ubiquitous in all examined genomes but with various gene number expansions. For example,Brassicaceaspecies contained many more JRLs (from 46 to 123) than the other 25 plants (from 2 to 41), indicating that the expansion of JRLs inBrassicaceaspecies has been faster than those in the other plants, such as moss, fern,and other angiosperms. InPoaceae, rice has a small size genome (380 Mb) but contains 30 JRL genes (latest version of MSU rice genome project) (Table 2), suggesting that riceJRLsgenes have experienced a faster gene expansion rate compared toB.distachyon,H.vulgare,S.bicolor, andZ.mays. For rice and maize, both of which have polyploid origin, whole-genome duplication occurred in the common rice and maize ancestor (at 70–80 mya (million years ago))followed by genome diploidization (including some loss of duplicated genes) before the divergence of rice-maize (at 50 mya) from the polyploid ancestor (Gautet al. 1997;Patersonet al. 2004; Wanget al. 2005; Llacaet al. 2011).The genome contraction most probably has continued further in the maize line and removed more duplicated genes, during at least two more polyploidization-contraction events (Patersonet al. 2004; Llacaet al. 2011), while in rice more of the originally duplicated genes were retained instead of diversifying to gain new functions. Ancient polyploidization and subsequent diploidization (loss) of many duplicated gene copies have shaped the genomes of allPoaceaecereal, forage, and biomass crops.
4.2. Domain combination of Poaceae JRLs
JRLs from 30 selective genomes are investigated with different combinations of jacalin and other domains. In dicots, the predominating types are different for species except in theBrassicaceaefamily. FiveBrassicaceaespecies encode for more holojacalins than the remaining two. Most of the holojacalins contain more than two identical jacalin domains. In other dicots species, the distributions of the major jacalin types are not associated with evolutionary relationships (Fig. 1), which may be due to the limited number of genomes we investigated. JRLs of dicots are different from that of monocots, but both of them have similarity with mosses. Monocot species have merojacalins as their major type, chimeric jacalins as the second most abundant, holojacalins as the third. Six holojacalins from five grass species contain more than two repeated jacalin domains. This result is different from what have been found by other authors (Songet al. 2014) and it may be due to updated versions of the selected grass genomes.Poaceaespecies contain a higher proportion of chimeric jacalins (40%) than dicots in this study. The jacalin domains in grass chimeric jacalins are covalently linked to dirigent, Pkinase, NB_ARC or B3 domain and so on. Two types of chimeric jacalins are specific to grass species (Fig. 3). One type is dirigent-jacalin domains found in all six grass species which further confirmed the results reported by Song (2014). Another type is NB_ARC-jacalin from wheat, sorghum, barley, and rice. Proteins with dirigent or disease-response domain exists in plants and have been implicated in plant defense (Ralphet al. 2007;Seneviratneet al. 2015; Weidenbachet al. 2016). Jacalins proteins with N-terminal dirigent domains have only been found inPoaceaespecies (Songet al. 2014) but we have now confirmed this by a more detailed study. RiceOsJAC1(Os12g14440) gene is encoded with aPoaceae-specific jacalin-related lectin and is required for resistance against rice pathogens by containing dirigent and jacalin domains(Jianget al. 2006; Weidenbachet al. 2016). In wheat,dirigent-JRL TaHfr-1 (Hessian fly-responsegene1) shows a positive transcription response to virulent Hessian fly larvae infection (Williamset al. 2002). Two dirigent-lead jacalins in wheat TaJRLL1 and TaJA1 (Traes_2BS_A1F541056.1)both confer enhanced resistance against pathogenic fungi in wheat immunity (Wang and Ma 2005; Xianget al. 2011;Maet al. 2013). Many plant resistance genes contain NBARC (nucleotide-binding proteins involved in ubiquitination)domains that regulate defense responses (De Schutter and Van Damme 2015). In this study, eight of the NB_ARC-jacalins are only found in grass species. It is possible that NB_ARC domain in JRLs protect grass species from pathogen attacks. From monocots to dicots, the proportion of chimeric jacalins increase while holojacalins decrease, it is possible that JRLs with multiple copies of jacalin domains will not fuse to other domains and be more prone to be lost during evolution to acquire new functions as JRLs with single domains.
In rice,JRLsgenes are distributed over nine out of 12 chromosomes. SixJRLsclusters with 27 genes in total were found in the rice genome. Ten rice chimeric jacalins encoded with NB_ARC, Pkinase or dirigent domain are found in a cluster on chromosomes 11 and 12. Three pairs of these 10 JRLs were present as monophyletic groups. Since rice diverged from wheat-barley much later than that from maize lineage 50 mya (Kellogg 2001), newly evolved rice monophyletic pairs may have occurred after the divergence of rice from wheat-barley or maize-sorghum.
4.3. Poaceae JRLs genes in response to pathogens
Some already knownJRLsgenes that encode with merojacalins or chimeric jacalins are regulated by biotic or abiotic stresses. For example, rice merojacalinSalTand chimeric jacalinOsJAC1both show positive responses to several stress treatments, including salt, drought, cold,salicylic acid (SA), methyl jasmonate (Me-JA), and abscisic acid (ABA) (Claeset al. 1990; Garciaet al. 1998; De Souza Filhoaet al. 2003; Jianget al. 2006; Weidenbachet al.2016). In wheat,JRLsshow different response patterns toward abiotic agents suggesting thatTaJRLsmay have complicated roles in plant defense response (Songet al.2014). In the present study, we examined the transcripts of riceJRLsgene in response to rice blast disease. Most of the riceJRLs(23) show transcriptional responses toM.oryzaeinoculations while the remaining (7) was not responsive.Some of these nonresponsive genes were previously also not found to respond toM.oryzae(Bagnaresiet al. 2012).These genes can be expressedin other tissues or under other environmental conditions. The expression patterns ofOsJRLs are basically the same in response to bothM.oryzaeGuy11 and FJ81278 strains, although it appears like that rice reacts slower to FJ81278 than to Guy11. Most of the responsive genes are up-regulated at specific stages of infections, such asOsJRLsgenes in groups 1, 2, 4,and 6 (Fig. 5). SomeOsJRLsgenes (groups 3 and 5) are suppressed afterM.oryzaeinoculation, especially at the early infection stage (Group 3 at 24 hpi).
Rice JRLs are largely characterized by structure diversity and pathogen inducible-expression, which also applies to other plants, such as wheat (Songet al. 2014).Poaceaehave evolved uniqueJRLsgenes by recruiting exogenous domains for certain adaptations. All detectedPoaceaespecificOsJRLgenes responded toM.oryzaeinoculation in this study (Fig. 5). The detailed functions of these uniqueJRLsgenes should be investigated in future studies.
5. Conclusion
We identified 651 jacalin-related lectins and analyzed the evolutionary trajectory and domain diversification in 30 plants species. We found the number and domain combination of JRLs proteins are highly variable in those plants.Poaceaespecies have evolved somePoaceae-specific JRLs. RiceJRLsgenes displayed variable transcriptional changes uponM.oryzaeinfections.
Acknowledgements
We are graceful to MSc Lin Lianyu of Fujian Agriculture and Forestry University for assistance in sequencing result analysis. This project is funded by the National Key Research and Development Program of China(2016YFD0100600) and the National Natural Science Foundation of China (U1405212).
Appendicesassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Abebe T, Skadsen R, Patel M, Kaeppler H. 2006. TheLem2gene promoter of barley directs cell- and developmentspecific expression ofgfpin transgenic plants.Plant Biotechnology Journal,4, 35–44.
Altschul S F, Madden T L, Sch?ffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. 1997. Gapped BLAST and PSI-BLAST:A new generation of protein database search programs.Nucleic Acids Research,17, 3389–3402.
Bagnaresi P, Biselli C, Orrù L, Urso S, Crispino L, Abbruscato P, Piffanelli P, Lupott, E, Cattivelli L, Valè G. 2012.Comparative transcriptome profiling of the early response toMagnaporthe oryzaein durable resistantvs. susceptible rice (Oryza sativaL.) genotypes.PLoS ONE,7, e51609.
Blanchard D, Cicek M, Chen J, Esen A. 2001. Identification of beta-glucosid- ase aggregating factor (BGAF) and mapping of BGAF binding regions on maize beta-glucosidase.Journal of Biological Chemistry,276, 11895–11901.
Bourne Y, Roig-Zamboni V, Barre A, Peumans W J, Astoul C H, Van Damme E J, Rougé P. 2004. The crystal structure of the Calystegia sepium agglutinin reveals a novel quaternary arrangement of lectin subunits with a beta-prism fold.Journal of Biological Chemistry,279, 527–533.
Bourne Y, Zamboni V, Barre A, Peumans W J, Van Damme E J, Rougé P. 1999. Helianthus tuberosus lectin reveals a widespread scaffold for mannosebinding lectins.Structure,7, 1473–1482.
Chang W C, Liu K L, Hsu F C, Jeng S T, Cheng Y S. 2012.Ipomoelin, a jacalin-related lectin with a compact tetrameric association and versatile carbohydrate binding properties regulated by its N terminus.PLoS ONE,7, e40618.
Chen J, Zheng W, Zheng S, Zhang D, Sang W, Chen X, Li G,Lu G, Wang Z. 2008. Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogenMagnaporthe grisea.PLoS Pathogens,4, e1000202.
Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G,Van Montagu M, Caplan A. 1990. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought.The Plant Cell,2, 19–27.
Van Damme E J, Barre A, Mazard A M, Verhaert P, Horman A,Debray H, Rougé P, Peumans W J. 1999. Characterization and molecular cloning of the lectin fromHelianthus tuberosus.European Journal of Biochemistry,259,135–142.
Van Damme E J, Barre A, Verhaert P, Rougé P, Peumans W J. 1996. Molecular cloning of the mitogenic mannose/maltose-specific rhizome lectin fromCalystegia sepium.FEBS Letter,397, 352–356.
Van Damme E J, Hause B, Hu J, Barre A, Rougé P, Proost P, Peumans W J. 2002. Two distinct jacalin-related lectins with a different specificity and subcellular location are major vegetative storage proteins in the bark of the black mulberry tree.Plant Physiology,130, 757–769.
Van Damme E J, Zhang W, Peumans W J. 2004. Induction of cytoplasmic mannose-binding jacalin-related lectins is a common phenomenon in cereals treated with jasmonate methylester.Communications in Agricultural and Applied Biological Sciences,69, 23–31.
Esen A, Blanchard D J. 2000. A specific beta-glucosidaseaggregating factor is responsible for the beta-glucosidase null phenotype in maize.Plant Physiology,2, 563–572.
Feng H, Xu W Z, Lin H H, Chong K. 2009. Transcriptional regulation of wheat VER2 promoter in rice in response to abscisic acid, jasmonate, and light.Journal of Genetics and Genomics,6, 371–377.
Garcia A B, Engler Jde A, Claes B, Villarroel R, Van Montagu M, Gerats T, Caplan A. 1998. The expression of the saltresponsive gene salT from rice is regulated by hormonal and developmental cues.Planta,2, 172–180.
Gaut B S, Doebley J F. 1997. DNA sequence evidence for the segmental allotetraploid origin of maize.Proceedings of the National Academy of Sciences of the United States of America,13, 6809–6814.
G?rlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U,Kogel K H, Oostendorp M, Staub T, Ward E, Kessmann H,Ryals J.1996. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat.The Plant Cell,4, 629–643.
Grunwald I, Heinig I, Thole H H, Neumann D, Kahmann U, Kloppstech K, Gau A E. 2007. Purification and characterisation of a jacalin-related, coleoptile specific lectin fromHordeum vulgare.Planta,226, 225–234.
Hagel J M, Facchini P J. 2010. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy.Nature Chemical Biology,4, 273–275.
Hosmani P S, Kamiya T, Danku J, Naseer S, Geldner N,Guerinot M L, Salt D E.2013. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-basedCasparian stripin the root.Proceedings of the National Academy of Sciences of the United States of America,35, 14498–14503.
Imanishi S, Kito-Nakamura K, Matsuoka K, Morikami A,Nakamura K. 1997. A major jasmonate-inducible protein of sweet potato, ipomoelin, is an ABA-independent woundinducible protein.Plant Cell Physiology,38, 643–652.
Jiang J F, Han Y, Xing L J, Xu Y Y, Xu Z H, Chong K. 2006.Cloning and expression of a novel cDNA encoding a mannose-specific jacalin-related lectin fromOryza sativa.Toxicon,1, 133–139.
Jiang S Y, Ma Z, Ramachandran S. 2010. Evolutionary history and stress regulation of the lectin superfamily in higher plants.BMC Evolutionary Biology,10, 79.
Katoh K, Standley D M. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability.Molecular Biology and Evolution,30, 772–780.
Kellogg E A. 2001. Evolutionary history of the grasses.Plant Physiology,3, 1198–1205.
Kittur F S, Lalgondar M, Yu H Y, Bevan D R, Esen A. 2007.Maize beta- glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for beta-glucosidase aggregation.Journal ofBiological Chemistry,10, 7299–7311.
Li B, Dewey C N. 2011. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome.BMC Bioinformatics,12, 323.
Lin J S, Lin H H, Li Y C, King Y C, Sung R J, Kuo Y W, Lin C C,Shen Y H, Jeng S T. 2014. Carbon monoxide regulates the expression of the wound-inducible gene ipomoelin through antioxidation and MAPK phosphorylation in sweet potato.Journal of Experimental Botany,18, 5279–5290.
Liu K, Linder C R, Warnow T. 2011. RAxML and FastTree:Comparing two methods for large-scale maximum likelihood phylogeny estimation.PLoS ONE,6, e27731.
Liu X Y, Li H, Zhang W. 2014. The lectin fromMusa paradisiacabinds with the capsid protein of tobacco mosaic virus and prevents viral infection.Biotechnology & Biotechnological Equipment,3, 408–416.
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta DeltaC (t)) method.Methods,4, 402–408.
Llaca V, Campbell M A, Deschamps S. 2011. Genome diversity in maize.Journal of Botany, doi: 10.1155/2011/104172
Ma Q H, Tian B, Li Y L. 2010. Overexpression of a wheat jasmonate-regulated lectin increases pathogen resistance.Biochimie,2, 187–193.
Ma Q H, Zhen W B, Liu Y C. 2013. Jacalin domain in wheat jasmonate-regulated protein Ta-JA1 confers agglutinating activity and pathogen resistance.Biochimie,2, 359–365.
Mistry J, Finn R D, Eddy S R, Bateman A, Punta M. 2013.Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions.Nucleic Acids Research,41, e121.
Moreira R A, Ainouz I L. 1981. Lectins from seeds of jack fruit(Artocarpus integrifoliaL.): Isolation and purification of two isolectins from the albumin fraction.European Journal of Biochemistry,23, 186–192.
Nagano A J, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I. 2008. Antagonistic jacalin-related lectins regulate the size of ER body-type beta-glucosidase complexes inArabidopsis thaliana.Plant and Cell Physiology,6, 969–980.
Nejat N, Mantri N. 2017. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses.Critical Reviews in Biotechnology,20, 1–13.
Paterson A H, Bowers J E, Chapman B A. 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics.Proceedings of the National Academy of Sciences of the United States of America,26, 9903–9908.
Peumans W J, Van Damme E J. 1995. Lectins as plant defense proteins.Plant Physiology,2, 347–352.
Peumans W J, Van Damme E J. 1995. The role of lectins in plant defence.The Histochemical Journal,4, 253–271.
Peumans W J, Van Damme E J, Barre A, Rougé P. 2001.Classification of plant lectins in families of structurally and evolutionary related proteins.Advances in Experimental Medicine and Biology,491, 27–54.
Peumans W J, Zhang W, Barre A, Astoul C H, Balint-Kurti P J,Rovira P, Rougé P, May G D, Van Leuven F, Truffa-Bachi P, Van Damme E J. 2000. Fruit-specific lectins from banana and plantain.Planta,211, 546–554.
Price M N, Dehal P S, Arkin A P. 2009. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix.Molecular Biology and Evolution,26,1641–1650.
Richly E, Kurth J, Leister D. 2002. Mode of amplification and reorganization of resistance genes during recentArabidopsis thalianaevolution.Molecular Biology and Evolution,1, 76–84.
Poiroux G, Pitié M, Culerrier R, Ségui B, Van Damme E J,Peumans W J, Bernadou J , Levade T, Rougé P, Barre A, Benoist H. 2011. Morniga G: A plant lectin as an endocytic ligand for photosensitizer molecule targeting toward tumorassociated T/Tn antigens.Photochemistry and Photobiology,2, 370–377.
Ralph S G, Jancsik S, Bohlmann J. 2007. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny,and constitutive and stress-induced gene expression in spruce (Picea spp.).Phytochemistry,14, 1975–1991.
Sandalio L M, Romero-Puertas M C. 2015. Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signaling networks.Annals of Botany,4, 475–485.
Sahay S, Gupta M. 2017. An update on nitric oxide and its benign role in plant responses under metal stress.Nitric Oxide,67, 39–52.
Schmittgen T D, Livak K J. 2008. Analyzing real-time PCR data by the comparative C(T) method.Nature Protocols,6, 1101–1108.
De Schutter K, Van Damme E J. 2015. Protein-carbohydrate interactions as part of plant defense and animal immunity.Molecules,20, 9029–9053.
Seneviratne H K, Dalisay D S, Kim K W, Moinuddin S G, Yang H, Hartshorn C M, Davin L B, Lewis N G.2015. Non-host disease resistance response in pea (Pisumsativum)pods:Biochemical function of DRR206 and phytoalexin pathway localization.Phytochemistry,113, 140–148.
Song M, Xu W, Xiang Y, Jia H, Zhang L, Ma Z. 2014. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling.Plant Molecular Biology,84, 95–110.
De Souza Filhoa G A, Ferreiraa B S, Diasa, M J, Queiroza K S,Brancoa A T, Bressan-Smithb R E, Oliveirab J G, Garciaa A B. 2003. Accumulation of SALT protein in rice plants as a response to environmental stresses.Plant Science,164,623–628.
Tateno H, Winter H C, Petryniak J, Goldstein I J. 2003.Purification, characterization, molecular cloning, and expression of novel members of jacalin-related lectins from rhizomes of the true fernPhlebodium aureum(L.) J.Smith (Polypodiaceae).Journal of Biological Chemistry,278, 10891–10899.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley D R, Pimentel H, Salzberg S L, Rinn J L, Pachter L. 2012.Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks.Nature Protocols,7, 562–578.
Vandenborre G, Smagghe G, Van Damme E J. 2011. Plant lectins as defense proteins against phytophagous insects.Phytochemistry,13, 1538–1550.
Vandenborre G, Smagghe G, Van Emms D M, Kelly S. 2015.OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy.Genome Biology,16, 157.
Wang X, Shi X, Hao B, Ge S, Luo J. 2005. Duplication and DNA segmental loss in the rice genome: Implications for diploidization.New Phytologist,3, 937–946.
Wang X M, Ma Q H. 2005. Characterization of a jasmonateregulated wheat protein related to a beta-glucosidaseaggregating factor.Plant Physiology and Biochemistry,2,185–192.
Weidenbach D, Esch L, M?ller C, Hensel G, Kumlehn J, H?fle C, Hückelhoven R, Schaffrath U. 2016. Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modularPoaceae-specific proteins.Molecular Plant,4, 514–527.
Williams C E, Collier C C, Nemacheck J A, Liang C, Cambron S E. 2002. A lectin-like wheat gene responds systemically to attempted feeding by avirulent first-instar Hessian fly larvae.Journal of Chemical Ecology,7, 1411–1428.
Xiang Y, Song M, Wei Z, Tong J, Zhang L, Xiao L, Ma Z,Wang Y. 2011. A jacalin-related lectin-like gene in wheat is a component of the plant defence system.Journal of Experimental Botany,15, 5471–5483.
Xiao J, Li C, Xu S, Xing L, Xu Y, Chong K. 2015. JACALINLECTIN LIKE1 regulates the nuclear accumulation of GLYCINE-RICH RNA-BINDING PROTEIN7, influencing the RNA processing ofFLOWERING LOCUS Cantisense transcripts and flowering time inArabidopsis.Plant Physiology,3, 2102–2117.
Xing L, Li J, Xu Y, Xu Z, Chong K. 2009. Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility.PLoS ONE,3, e4854.
Yamaji Y, Maejima K, Ozeki J, Komatsu K, Shiraishi T, Okano Y, Himeno M, Sugawara K, Neriya Y, Minato N, Miura C,Hashimoto M, Namba S.2012. Lectin-mediated resistance impairs plant virus infection at the cellular level.The Plant Cell,2, 778–793.
Yong W D, Xu Y Y, Xu W Z, Wang X, Li N, Wu J S, Liang T B,Chong K, Xu Z H, Tan K H, Zhu Z Q.2003. Vernalizationinduced flowering in wheat is mediated by a lectin-like geneVER2.Planta,2, 261–270.
 Journal of Integrative Agriculture2018年6期
Journal of Integrative Agriculture2018年6期
- Journal of Integrative Agriculture的其它文章
- Improve access to the EU market by identifying French consumer preference for fresh fruit from China
- Management and prevention of mastitis: A multifactorial approach with a focus on milking, bedding and data-management
- Elimination of ceftiofur hydrochloride residue in postpartum cows’milk after intramammary infusing at dry-off
- Evaluation of a new qPCR test to identify the organisms causing high total bacterial count in bulk tank milk
- Prevalence and characteristics of extended spectrum β-lactamaseproducing Escherichia coli from bovine mastitis cases in China
- Molecular mapping of YrTZ2, a stripe rust resistance gene in wild emmer accession TZ-2 and its comparative analyses with Aegilops tauschii
