Occurrence of K1 and K2 serotypes and genotypic characteristics of extended spectrum β-lactamases-producing Klebsiella pneumoniae isolated from selected hospitals in Malaysia
Nurul Syazrah Anuar ,Hazmin Hazman ,Sharven Raj Jeyakumar ,Mohd Nasir Mohd Desa ,Hasni Idayu Saidi ,Siti Norbaya Masri ,Nur Afiza Aziz,Nurshahira Sulaiman?
1Department of Biomedical Science,Faculty of Medicine and Health Sciences,Universiti Putra Malaysia,43400,UPM Serdang,Selangor,Malaysia
2Department of Medical Microbiology,Faculty of Medicine and Health Sciences,Universiti Putra Malaysia,43400,UPM Serdang,Selangor,Malaysia
3Microbiology Unit,Department of Pathology,Hospital Sultanah Aminah,Jalan Persiaran Sultan Abu Bakar,80100 Johor Bahru,Johor,Malaysia
ABSTRACT Objective:To determine the distribution,phenotypic and genetic background of extended spectrum β-lactamases (ESBL)-producing Klebsiella (K.) pneumoniae clinical isolates associated with K1 and K2 serotypes in two selected hospitals in Malaysia.Methods:A total of 192 K.pneumoniae isolates were collected and subjected to antibiotic susceptibility,hypermucoviscosity test and multiplex PCR to detect the presence of K1-and K2-serotype associated genes.Multilocus sequence typing (MLST) was performed on ESBL-producing K.pneumoniae isolates presented with K1 and K2 serotypes,followed by phylogenetic analysis.Results:A total of 87 out of 192 (45.3%) of the K.pneumoniae isolates collected were ESBL producers.However,only 8.3%(16/192) and 10.9% (21/192) of the total isolates were detected to carry K1-and K2-serotype associated genes,respectively.Statistical analysis showed that K1 and K2 capsular serotypes were not significantly associated with ESBL phenotype (P=0.196).However,they were significantly associated with hypervirulent,as demonstrated by the positive string test (P<0.001).MLST analysis revealed that ST23 as the predominant sequence type (ST) in the K1 serotype,while the ST in the K2 serotype is more diverse.Conclusions:Although the occurrence of ESBL-producing isolates among the hypervirulent strains was low,their coexistence warrants the need for continuous surveillance.MLST showed that these isolates were genetically heterogeneous.
KEYWORDS: Extended spectrum β-lactamases;Klebsiella pneumoniae;Capsular serotypes;Genotypic
1.Introduction
Klebsiella (K.) pneumonia is responsible for many severe nosocomial infections such as pneumonia,urinary tract infections,bacteremia,meningitis,liver abscesses and endophthalmitis[1-3].Over the past few decades,these bacteria have been reported globally to display an increasing resistance profile towards β-lactamases antibiotics through the production of β-lactamase,such as extended-spectrum β lactamase (ESBL)[4,5].
Extended-spectrum β-lactamase is a plasmid-mediated enzyme that confers the ability to hydrolyze the β-lactam binding of β-lactam antibiotics,rendering them inactive[4].Recent studies have shown that ESBL-producing K.pneumoniae is resistant to the first,second,third and fourth generation cephalosporins,aztreonam and aminopenicillins but is inhibited by clavulanate[5].This is in line with the National Antibiotic Surveillance Report,2021 in Malaysia where an increasing resistance trend of K.pneumoniae to all antibiotics was observed[6].This poses a major therapeutic challenge in the treatment of infection as the range of treatment options is limited.
Overall,the occurrence of ESBL in K.pneumoniae varies widely among distinct geographic regions.Southeast Asia is considered the epicentre of ESBL-producing Enterobacteriaceae,and high rates of this phenotype,particularly among K.pneumoniae isolates[7,8].Since the occurrence of ESBL strains varies geographically,different regions reported considerably different occurrence and resistance patterns of ESBL-producing K.pneumoniae.Previous studies conducted in several Southeast Asian countries,including Laos,Thailand and Indonesia reported that ESBL production in K.pneumoniae ranged from 4.7% to 53%[1,7,8].
Over the years,K.pneumoniae has been classified according to its capsular type,which to date consists of 78 serotypes[9].Strains belonging to capsular serotypes K1 and K2 are known to have the highest virulent activity in humans compared to their counterparts of other serotypes[10,11].Sporadic cases of severe invasive infections have been reported involving K.pneumoniae strains with hypervirulence that were commonly associated with capsular serotypes K1 or K2[12,13].Several studies conducted in Spain,Singapore and China found that the high mortality rate in liver abscesses with bacteremia caused by K.pneumoniae of the capsular serotype K1 and K2[3,12,13].However,K1 and K2 capsular serotypes should not be used to solely define hypervirulence as there are variety of other virulence factors that might contribute to this condition[14].
Meanwhile,since magA and K2A genes are restricted to K1 and K2 capsular serotypes respectively[11],they serve as useful markers to identify the two capsular serotypes.Nevertheless,the information on the genotypic characteristics and the occurrence of K1 and K2 capsular serotypes in K.pneumoniae infection in Malaysia are still limited to provide a decent insight on the scenario of infection pattern related to K.pneumoniae.Therefore,a collection of clinical K.pneumoniae isolates from hospital settings were characterized for their demographic,antibiotic susceptibility,serotypes and genotypic characteristics.
2.Subjects and methods
2.1.Ethical approval
Ethical approval for this study was obtained from the Medical Research and Ethics Committee of the Ministry of Health Malaysia,the National Medical Research Register (NMRR-20-3087-57426)and the Medical Research Ethics Committee of Universiti Putra Malaysia (JKEUPM-2021-057).
2.2.Bacterial isolates
A total of 192 K.pneumoniae clinical isolates were included in this cross-sectional study.All isolates were collected from the Microbiology Unit of Hospital Sultanah Aminah,Johor Bahru(HSAJB) and Hospital Sultan Abdul Aziz Shah (HSAAS) between March 2021 to April 2022.The criteria for selecting the samples were non-duplicated clinical isolates identified as K.pneumoniae.HSAJB and HSAAS are tertiary hospitals serving the population in the respective areas in peninsular Malaysia.Isolates from both hospitals were analyzed altogether in relation to demographic,phenotypic and genotypic characteristics associated with the isolates for potential correlation.Whenever appropriate,Chi-square of Fisher's exact test was used in analyzing the categorical tabulation.
The K.pneumoniae isolates were confirmed by the following biochemical test characteristics: non-motile,indole negative,methyl red negative,oxidase test negative,Voges Proskauer test positive,citrate utilization test positive and catalase positive[15].Identification of hypermucoviscousity in K.pneumoniae isolates was confirmed by the string test[16].Colonies grown overnight on culture media were touched with a loop and stretched from the surface of the media to detect the formation of mucoid string.
2.3.Antibiotic susceptibility test and ESBL detection
The susceptibility of K.pneumoniae isolates to aztreonam (30 μg),ceftazidime (30 μg),ceftriaxone (30 μg) and cefotaxime (30 μg)Oxoid Ltd,Basigstokes UK was determined using the Kirby Bauer’s disc diffusion method according to the recommendations of the Clinical and Laboratory Standard Institute (CLSI,2022)[17].Isolates with a diameter of inhibition zone of ≤17 mm against ceftazidime,≤22 mm against aztreonam,≤27 mm against cefotaxime and ≤27 mm against ceftriaxone were suspected to be ESBL producers.K.pneumoniae ATCC 700603 was used as the quality control strain.
All isolates were then phenotypically screened to confirm ESBL production using ceftazidime (30 μg),ceftazidime-clavulanate (30/10μg),cefotaxime (30 μg),cefotaxime-clavulanate (30/10 μg) Oxoid Ltd,Basigstokes UK and Becton Dickinson,USA on Mueller-Hinton agar with inoculum 0.5 McFarland turbidity standard.After 16-18 hours of incubation at 37 ℃,the diameter of the inhibition zones was measured.An increase in zone diameter of 5 mm increase in zone diameter for either antimicrobial agent tested in combination with clavulanate compared to when the agent was tested alone was regarded as a ESBL producers according to the CLSI breakpoints 2022[17].
2.4.Serotyping by multiplex PCR
The detection of K1 and K2 serotypes of K.pneumoniae isolates was carried out by multiplex PCR method adapted from Al-Jailawi et al.2014[18] with slight modifications in the optimization according to Elnifro et al.2020[19].The primers used in this study were magA[20],K2A[21] and 16s rRNA[22].The amplification of magA and K2A genes was carried out to identify K1 and K2 capsular serotype among isolates,while 16s rRNA gene served as an internal control for the identification of K.pneumoniae isolates at species level.Subsequently,extracted DNA consisting the targeted fragments of magA,K2A and 16s rRNA genes were amplified using a BioRadMyCycler? Thermal Cycler (BioRad,USA).Non-typable isolates in this study were defined by the presence of 16s rRNA and the absence of magA and K2A genes.
2.5.Multilocus sequence typing (MLST)
Twelve isolates that were ESBL producers and positive for capsular serotype K1 or K2 were subjected to MLST.MLST was performed based on an existing scheme for K.pneumoniae targeting seven loci,including gapA,infB,mdh,pgi,phoE,rpoB and tonB[23].PCR amplification was performed using a BioRadMyCycler? Thermal Cycler (BioRad,USA) in a 50 μL reaction mixture.
The amplified seven housekeeping genes were sequenced at a commercial facility (1st Base Sequencing,Apical Scientific,Malaysia).Subsequently,DNA alignment was carried out using Molecular Evolutionary Genetic Analysis version 11 (MEGA11) for all seven gene loci sequences of the isolates including ten reference sequences available in GenBank,National Center for Biotechnology Information (NCBI).The allele number for the specific aligned sequences was obtained from the Pasteur Institute MLST databases(https://bigsdb.pasteur.fr/).Isolates were assigned according to their respective sequence type (ST) based on allele combinations in all seven loci.
2.6.Phylogenetic analysis
Phylogenetic analysis of the sequenced DNA of the MLST genes was also performed using MEGA software version 11[24],which is available on the website www.megasoftware.net.Maximum likelihood method was used based on the Tamura and Nei model[25]with bootstrap sampling at 1 000 replicates.The available references ID:62,ID:723,ID:847,ID: 942,ID:968,ID:2731,ID: 2998,ID:3017,ID:3900,ID:4799 and ID: 14156 for the respective STs were retrieved from the MLST database and used as controls in the analysis.
2.7.Statistical analysis
The data were statistically analyzed using the Statistical Package for Social Sciences (SPSS) software version 25.0 for Microsoft Windows (Chicago,USA).Descriptive statistics (frequency and percentage) are used to summarize the socio-demographic characteristics,antimicrobial susceptibility patterns and ESBL distribution.The Chi-square test was used to compare the association between serotypes,ESBL production and other variables among the K.pneumoniae isolates.The P-value at <0.05 indicates statistical significance.
3.Results
3.1.Demographic characteristics of patients infected with K.pneumoniae and their association with ESBL expression
Among the 192 K.pneumoniae isolates,109 (56.8%) and 83(43.2%) were isolated from male and female patients,respectively.As Malaysia is a multi-racial country,our isolates were isolated from different ethnic groups;in which majority of them being Malays(n=132,68.8%),followed by Chinese (n=32,16.7%),Indians (n=19,9.9%) and others (n=9,4.7%).Meanwhile,the under-65 age group made up the majority of isolates (78%).The distribution of ESBL and non-ESBL K.pneumoniae isolates in relation to sex,ethnicity,age group and type of specimen is shown in Table 1.Chi-square analysis showed that sex and age group were not significantly associated with ESBL expression (P>0.05).On the other hand,statistically significant associations was observed between ESBL status and specimen type (P=0.045) (Fisher-Freeman-Halton Exact Test).
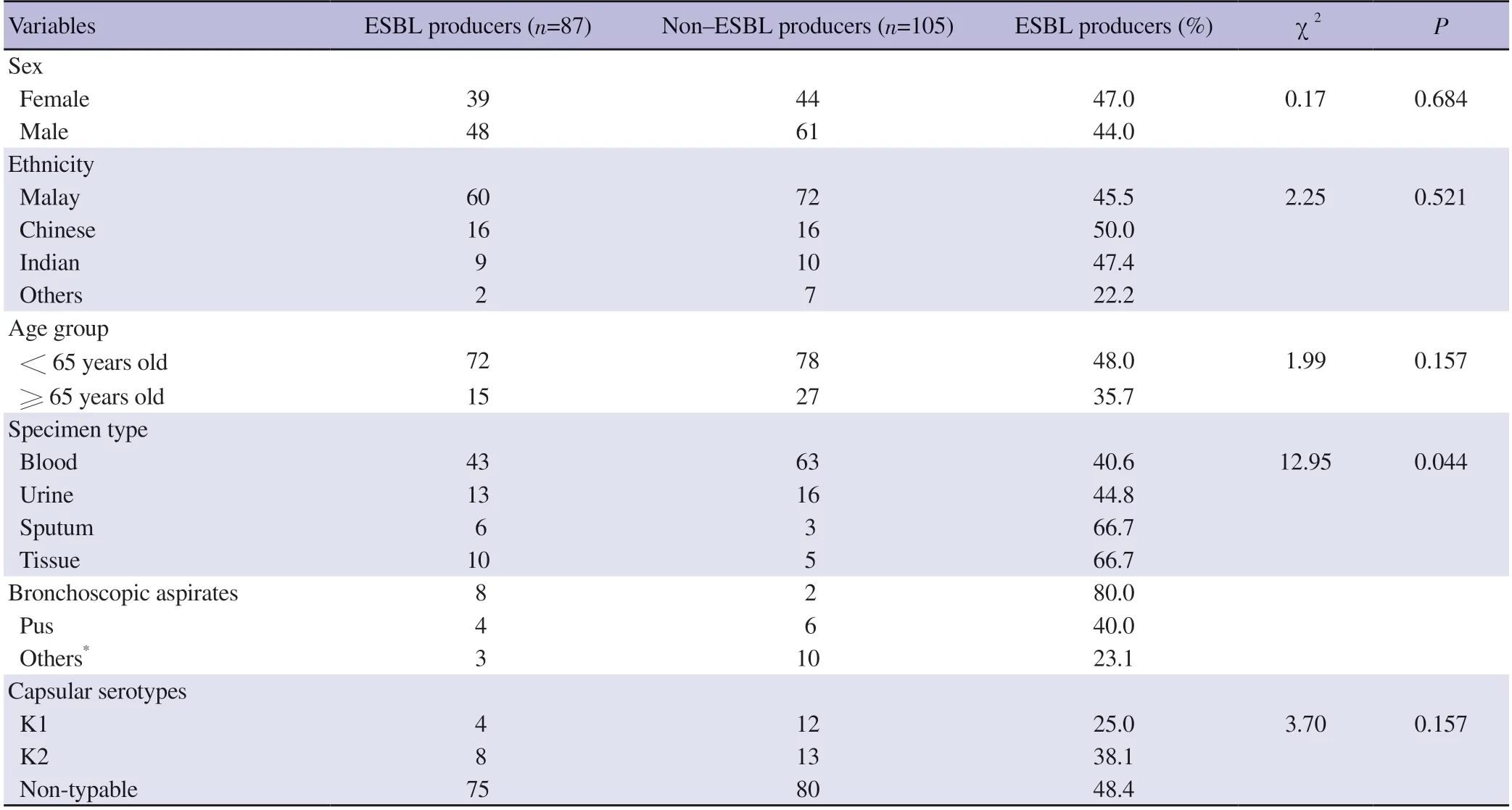
Table 1. Distribution of the ESBL and non-ESBL Klebsiella pneumoniae clinical isolates in relation to sex,ethnicity,age group and specimen type.
3.2.Antibiotic susceptibility test
As tabulated in Table 2,this study found that ESBL-producing isolates had significantly higher percentages of resistance compared with non-ESBL producing isolates against all the four antibiotics tested.The mean zones of inhibition of ESBL-producing isolates were two to four times lower than those of non-ESBL producing isolates against aztreonam,ceftriaxone,ceftazidime and cefotaxime(Supplementary Table 1).
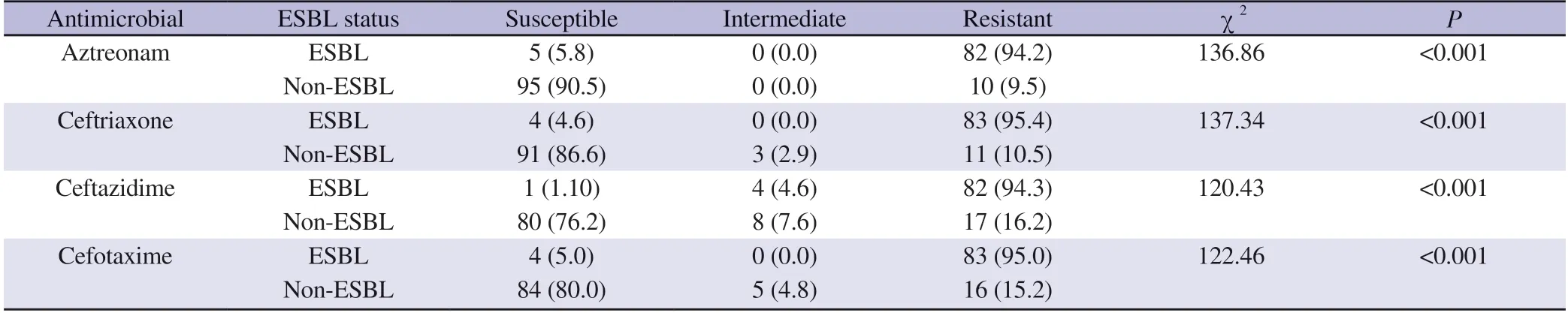
Table 2 .Susceptibility of the Klebsiella pneumoniae clinical isolates towards aztreonam,ceftriaxone,ceftazidime and cefotaxime [n (%)].
3.3.Detection of virulence factors in K.pneumoniae isolates
In the multiplex PCR,magA gene (1 283 bp) was detected in 16/192(8.3%) of the K.pneumoniae clinical isolates.The occurrence of the magA gene among ESBL and non-ESBL isolates was 4.6% (4/87)and 11.4% (12/105),respectively.On the other hand,K2A gene (543 bp) were detected in 21/192 (10.9%) isolates.The occurrence of the K2A gene was 9.2% (8/87) and 12.4% (13/105) in ESBL and non-ESBL strains,respectively (Supplementary Figure 1 shows the representative gel image of the detection).
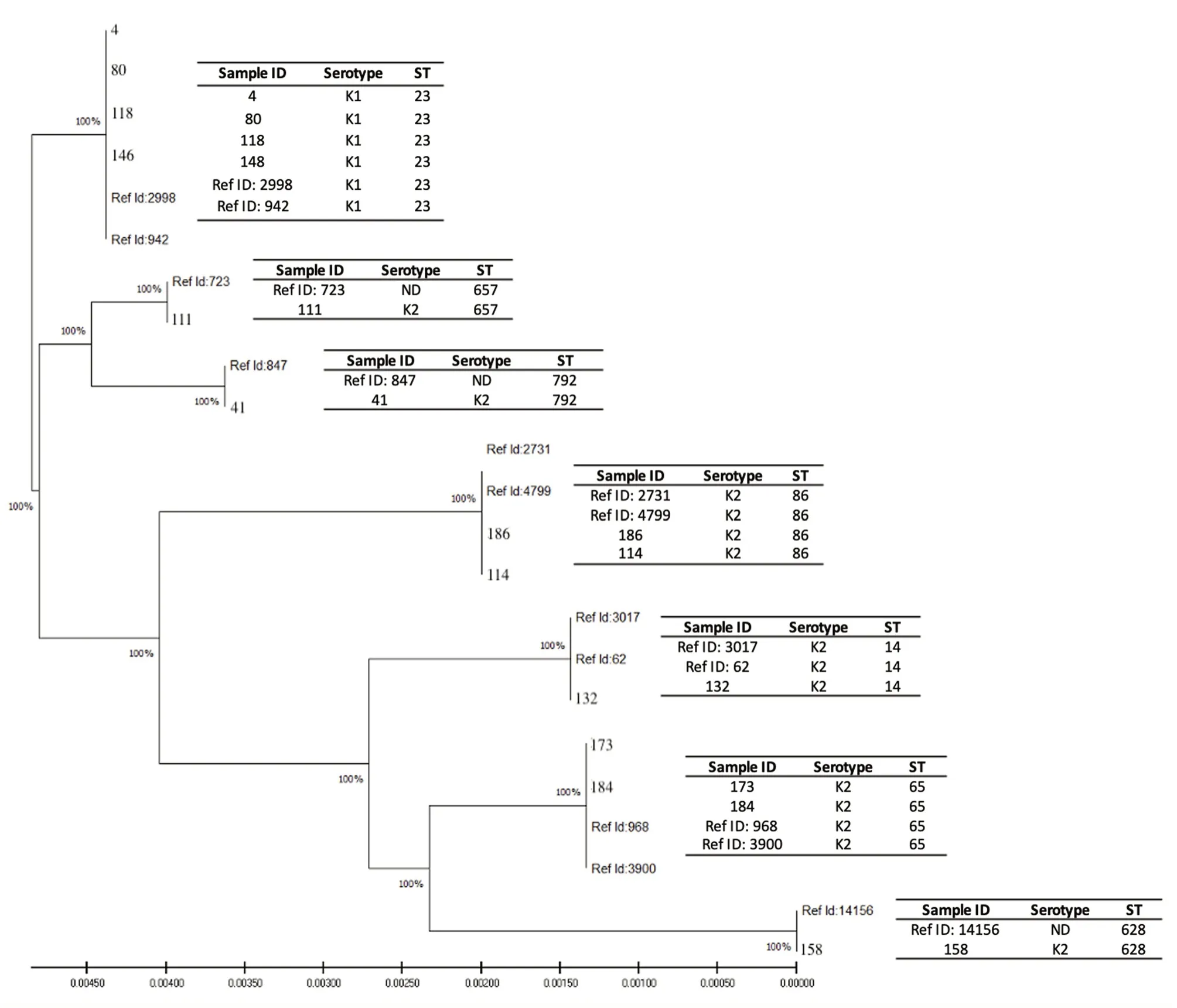
Figure 1.Phylogenetic analysis showed two distinct clusters,Clades A and B,with 100% bootstrap values at all branching.The distribution of serotype and sequence type (ST) for respective isolates are shown in the right column.Reference sequences were retrieved from the MLST database comprising identity numbers (ID) 2998 (Singapore),942 (Vietnam),723 (China),847 (Netherland),2731 (Guadeloupe,France),4799 (Moscow,Russia),3017 (Vietnam),62(Curacao),968 (Vietnam) and 3900 (Moscow,Russia).Numbers represent the Sample ID of the isolates in this study.
3.4.Determination of hypermucoviscousity phenotype
As shown in Table 3,the string test showed that 54 out of the 192 isolates (28.1%) were conferred positive for the hypermucoviscousity phenotype.Of these,50% (n=27) of the isolates were K1 (n=13)and K2 (n=14) capsular serotypes,respectively.The remaining 50%of the isolates that tested positive in the string test were non-K1 or K2 serotypes.Meanwhile,among the 87 ESBL-producing isolates,only 19.5% (n=17) displayed positive string test.A higher occurrence which is 35.2% positive string test,was observed among the non-ESBL isolates.Chi-square test was performed which revealed a significant association between capsular serotypes and hypermucoviscosity (P<0.001) as well as between ESBL status and hypermucoviscosity (P=0.023).
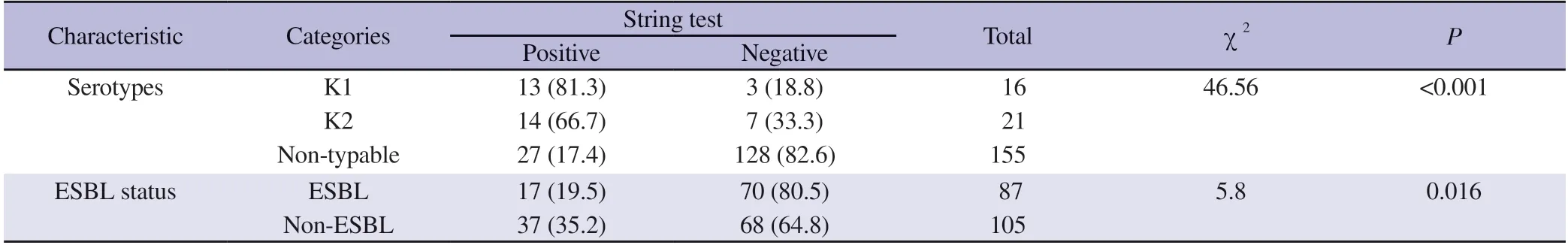
Table 3. String test observed in the Klebsiella pneumoniae clinical isolates [n (%)].
3.5.MLST profiles of K1 and K2 isolates presented with ESBL phenotype
A total of 12 isolates were confirmed through PCR and antibiotic susceptibility tests to possess ESBL and K1/K2 serotypes.MLST analysis carried out on these isolates (n=12) yielded seven distinct STs based on genetic variations on seven housekeeping genes(Figure 1).ST23 (n=4) was the predominant genotype belonging to K1 capsular serotype.Other strains with K2 serotypes belonged to ST86 (n=2),ST14 (n=1),ST65 (n=2),ST628 (n=1),ST657 (n=1)and ST792 (n=1) (Supplementary Table 1).
3.6.Phylogenetic analysis
The generated phylogenetic tree in Figure 1 shows two distinct clusters,Clades A and B,with 100% bootstrap values for all branching.Clade A consists of four isolates from our study presented with K1 capsular serotype with respective STs of two reference isolates retrieved from the MLST global database (ID: 942 and 2998).Interestingly,this cluster comprises only ST23 isolates (ID:4,80,118 and 146).Meanwhile,Clade B consists the remaining eight isolates of various STs presented with K2 capsular serotype with the respective STs of nine reference isolates retrieved from the MLST global database (ID: 62,723,847,968,2731,3017,3900 and 14156).
4.Discussion
This study noted a considerably high occurrence of ESBL producers (45.3%) among the K.pneumoniae clinical isolates collection.In previous studies conducted in Malaysia,the percentage of ESBL-producing K.pneumoniae ranged from 27.8% to 47%[26-28].The wide range could be due to different population of patients served,attributable to different hospitals location and time.In this study,descriptive demographic analysis showed a pattern on the occurrence of ESBL-producing strains to be slightly higher in males than females (56.8% vs.43.2%).Nirwati et al.[29] in their study involving 167 K.pneumoniae specimens from a hospital in Indonesia also reported that the K.pneumoniae infection rate was higher in males than females (64.07% vs.35.93%).The present study also showed that ESBL-producing K.pneumoniae was more common in the younger age group of under 65 years,than in patients aged 65 and over (37.5% vs.7.8%).Our findings were in line with studies conducted in Thailand and China where ESBL-producing K.pneumoniae was more common among children aged 0-4 years[7,30].The emergence of multidrug-resistant K.pneumoniae in young children especially to newborns are related to prematurity,low body weight,intrauterine infection and extensive use of antimicrobial drugs[7,30].Nevertheless,our study was limited in the history and demographic data of the patients to address the higher incidence in the patients of under 65 years,including that in the sex category and antibiotic susceptibility pattern to other antibiotics.
As for the antibiotic susceptibility pattern,the ESBL-producing K.pneumoniae study isolates showed a higher frequency of incidence of resistance to aztreonam,ceftriaxone,ceftazidime and cefotaxime compared with the non-ESBL-producing isolates.More than 90%of the ESBL-producing isolates displayed resistance against all antibiotics tested in this study (ceftriaxone: 95.40%,cefotaxime:95.40%,aztreonam: 94.25%,and ceftazidime: 94.25%).These results are comparable to those reported by Gharrah et al.[31] and Shin and Ko[32] where ESBL-producing isolates showed significantly higher frequency of incidence of resistance to most β-lactams compared to non-ESBL-producing isolates.The production of ESBL encoded by β-lactamase related genes such as SHV,TEM and CTX-M were responsible for enzymatic antibiotic inactivation and modification which lead to failure in the function of the antibiotics[33,34].
In the multiplex PCR analysis,a small proportion of the K.pneumoniae isolates were determined as capsular serotypes K1 and K2,(8.3% and 10.9% respectively).Of the two serotypes sought,serotype K2 (n=21) had a higher occurrence than serotype K1(n=16) serotype.This is consistent with the findings reported in a study from south India by Remya et al.in 2018[11] where a higher frequency of K2 capsular serotype compared to K1 capsular serotype was observed (2.16% vs.0.27%).Similarly,in a study conducted by Guo et al.in China[35],K2 was also found to be the most common serotype (42.9%,36/84).Strains with K1 or K2 capsular serotypes and ESBL phenotypes were generally reported to be commonly associated with hypervirulent phenotypes due to the extent of clinical manifestations of infection[11,36].Of the 192 isolates included in our study,54 (28.1%) tested positive for string test despite their capsular serotype and ESBL status.A high occurrence of 81.3% (n=13/16) and 66.7% (n=14/21) of isolates belonging to the K1 and K2 serotypes respectively,were tested positive for string test.However,17.4% of the isolates with non-K1 or K2 serotypes were also presented with a positive string test.This finding supports that hypervirulence should not be exclusively restricted to K1 or K2 serotypes,although they are most commonly associated with hypervirulent infection[11,14].Choby et al.in 2019[9] in their study also reported that aside from hypermucoviscousity,there are additional factors for hypervirulence of K.pneumoniae such as siderophores system,colibactin toxin and biofilms formation.Since a high proportion of capsular serotypes K1 and K2 was tested positive for hypermucoviscousity in the present study,their coexistence with ESBL production raises a major concern.The simultaneous presence of two virulence factors observed in this study which are K1/K2 serotypes and ESBL-producing phenotype,could lead to hypervirulent strains with greater antibiotics resistance that can cause severe infections and complicate disease management[37].Therefore,isolates positive for ESBL production and capsular serotype K1 or K2 were subjected to MLST testing to determine their genetic backgrounds.
Subsequently,MLST showed the predominant ST23 (n=4,33.33%)among the group of isolates with K1/K2 capsular serotypes and ESBL-producing phenotype.All isolates belonging to this ST share the same capsular serotype,K1.They were clustered into a single Clade A,indicating a high degree of clonality of the ST23 group.In previous studies carried out in China and Singapore,ST23 was reported to be predominant in the hypervirulent K.pneumoniae(hvKP) and K1 serotypes of K.pneumoniae[12,35].In contrast,the second clade (Clade B) contains isolates with K2 capsular serotypes that were found to belong to several distinct,unrelated sequence types,consisting of ST14,ST65,ST86,ST628,ST657 and ST729.This is in accordance with some other findings which reported that K2 strains of K.pneumoniae appear to belong to a wider range of ST groups[13,38].Meanwhile,looking at the tabulation of the ESBL producers in the phylogenetic tree,there was no obvious genetic association between ESBL producers of K.pneumoniae with K1 and K2 serotypes as they were segregated in different clusters.
This study has several limitations.Only two hospitals were included with only 36 isolates were collected from HSAAS compared to 156 from HSAJB.Therefore,all isolates in the study were analyzed as a whole collection regardless of the hospital origin.A larger sample size involving more hospitals from different regions might better represent the pattern of K.pneumoniae infection in Malaysia.In addition,this study lacks of information on the capsular serotype other than K1 or K2 and the limited number of virulence genes examined in this study.Additional virulence-associated genes such as peg-344,iroB,iucA,plasmid-borne prmpA and prmpA2[39]should be investigated to gain a better understanding of the virulence potential of K.pneumoniae.
All in all,the spread of ESBL-producing K.pneumoniae is a major problem and should be closely monitored.Infections caused by these strains are very difficult to treat and become even more challenging when inappropriate treatments are given.Since K1 and K2 capsular serotypes are most commonly associated with hypervirulent infections and complications,early detection may alert physicians and help them confirm appropriate therapy for patients infected with these strains.Further in-depth studies related to genetic analysis of the common virulence-associated genes would help in elucidating the molecular nature of the hypervirulent strains.
Conflict of interest statement
We declare that there is no conflict of interest.
Funding
This work was supported by the Ministry of Higher Education under the Fundamental Research Grant Scheme (FRGS/1/2021/SKK0/UPM/02/8) and the Universiti Putra Malaysia Research University Grant Scheme (GP/IPS/2021/9702000).
Acknowledgements
The authors would like to thank the Director General of Health Malaysia for the permission to publish this paper.We would also like to thank our collaborators at HSAJB and HSAAS for their assistance in the isolates collection.
Authors’ contributions
All authors have made substantial contributions to all of the following: the conception and design of the study,data collection,data analysis and interpretation,drafting the article or revising it critically for important intellectual content;and final approval of the version to be submitted.
 Asian Pacific Journal of Tropical Medicine2024年1期
Asian Pacific Journal of Tropical Medicine2024年1期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Unveiling health rights: A call to action for sex workers' HIV care in the Philippines
- Fatal cases in pediatric patients after post-exposure prophylaxis for rabies: A report of two cases
- Clinical profile and risk factors of Strongyloides stercoralis infection
- Informing policy makers in developing countries: Practices and limitations of geriatric home medication review in Malaysia-A qualitative inquiry
- Iron supplementation during malaria infection in pregnancy and childhood: A review
- Ferritin and mortality in hemodialysis patients with COVID-19: A systematic review and meta-analysis
