Development and application of KASP marker for high throughput detection of the seedless trait in grapevine
WANG Fu-qiang, BIAN Lu, QIU Peng-peng, GUO Shuo, GUO Jing-han, GUO Chen-shuo, JIANG Jian-fu, LIU Chong-huai, WANG Yong, LIU Guo-tian, WANG Yue-jin, XU Yan#
1 Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture and Rural Affairs/State Key Laboratory for Crop Stress Resistance and High-Efficiency Production/College of Horticulture, Northwest A&F University, Yangling 712100, P.R.China
2 Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou 450009, P.R.China
3 Research Institute of Grape and Melon of Xinjiang Uygur Autonomous Region, Shanshan 838200, P.R.China
Abstract Molecular marker-assisted selection (MAS) can significantly accelerate and improve the efficiency of the breeding process in seedless grape cultivars.In this study, we developed the KASP_VviAGL11 and VviAGL11_410 markers based on a single nucleotide polymorphism (SNP) site (Chr18: 26889437 (A/C)) of the VviAGL11 gene, and compared them with previously reported SSR markers p3_VvAGL11 and 5U_VviAGL11 by testing 101 cultivars and 81 F1 hybrid progenies.The results showed that both of the proposed markers obtained 100% accuracy rates in detecting allele A, which was closely associated with the seedless trait in grapes, while p3_VvAGL11 and 5U_VviAGL11 had lower accuracy rates due to their tendency to produce false positives.After careful evaluation of the technical advantages and disadvantages associated with these markers, we concluded that KASP_VviAGL11 was superior in terms of simplicity,cost-effectiveness, efficiency, and accuracy.Thus, we optimized the process of molecular MAS for seedless grapes,focusing on the KASP_VviAGL11 marker as a central component, to provide key technical support for the development of new seedless grape cultivars.
Keywords: seedless grape, MAS, KASP, SSR
1.Introduction
Grape (Vitis L.) is one of the most significant fruit crops worldwide.Seedless grape cultivars have become increasingly popular since they are preferred by both consumers and producers, making seedlessness a critical objective in grape breeding programs (Ledbetter and Ramming 1989; Varoquaux et al.2000).However, the development of these cultivars to produce new cultivars
faces many obstacles, including poor breeding efficiency,
high costs and lengthy breeding cycles.Therefore,accelerating the effectiveness of grape breeding is urgently needed.It is widely known that performing precise and efficient molecular marker-assisted selection(MAS) for seedless grapes during the seedling stage is vital for improving grape breeding efficiency and minimizing the associated costs (Bergamini et al.2013;Bennici et al.2019).
To date, various types of seedless molecular markers have been developed in grapevine, such as random amplified polymorphic DNA (RAPD) markers, sequence characterized amplified region (SCAR) markers, and simple sequence repeats (SSR) markers.Previously,the SCAR markers SCC8-1080 and SCF27-2000 were developed through bulk segregant analysis (BSA) by associating the imbalance with a seed development inhibitor (SDI), and they were found to be suitable for populations with both parents being seedless (Lahogue et al.1998; Mejía and Hinrichsen 2003).Subsequently,a RAPD marker named GSLP1-569 was developed with ‘Thompson Seedless’ as the test material (Wang and Olusola 2002), but this marker was only limited to‘Thompson Seedless’ and its closely related seedless cultivars (Li et al.2015, 2018).The VMC7f2 marker,located at the upstream 246 bp position of the 5′ UTR of the VviAGL11 (GSVIVT01025945001) gene (Pellerone et al.2001), was significantly associated with the seedless trait (Cabezas et al.2006; Royo et al.2018), but its identification efficiency varied significantly with the genetic background (Mejía et al.2011; Akkurt et al.2012).Mejía et al.(2011) developed an SSR marker, p3_VvAGL11,which is located in the 5′ UTR of the VviAGL11 gene with a (GAGA)nrepetitive sequence based on quantitative trait locus (QTL) mapping results (Fig.1).This marker had a low false positive rate in the germplasm across different genetic backgrounds, and the 198 bp allele was identified as closely associated with the seedless trait (Bergamini et al.2013).Furthermore, another SSR marker, 5U_VviAGL11 (Fig.1), was developed from the same region as p3_VvAGL11, with a longer amplification band, and the 319 bp allele was found to be significantly associated with the seedless trait.Chen et al.(2021) evaluated the SCF27-2000, GSLP1-569, VMC7f2, p3_VvAGL11,and 5U_VviAGL11 markers in 96 cultivars and 87 hybrid progenies, and concluded that the SSR markers p3_VvAGL11 and 5U_VviAGL11 were the most accurate and efficient in identifying the seedless germplasm.However,there are inconsistencies among studies in identifying the alleles closely associated with the seedless trait using SSR markers, making data comparison difficult.For example, while Mejía et al.(2011) identified the 198 bp allele amplified by the p3_VvAGL11 marker as closely associated with the seedless trait, Bennici et al.(2019),Ocarez et al.(2020), and Chen et al.(2021) respectively identified the 194, 196 and 187 bp alleles amplified by thep3_VvAGL11 marker in this respect.This discrepancy may be caused by systematic errors resulting from the use of different dye reagents and instruments for detection.
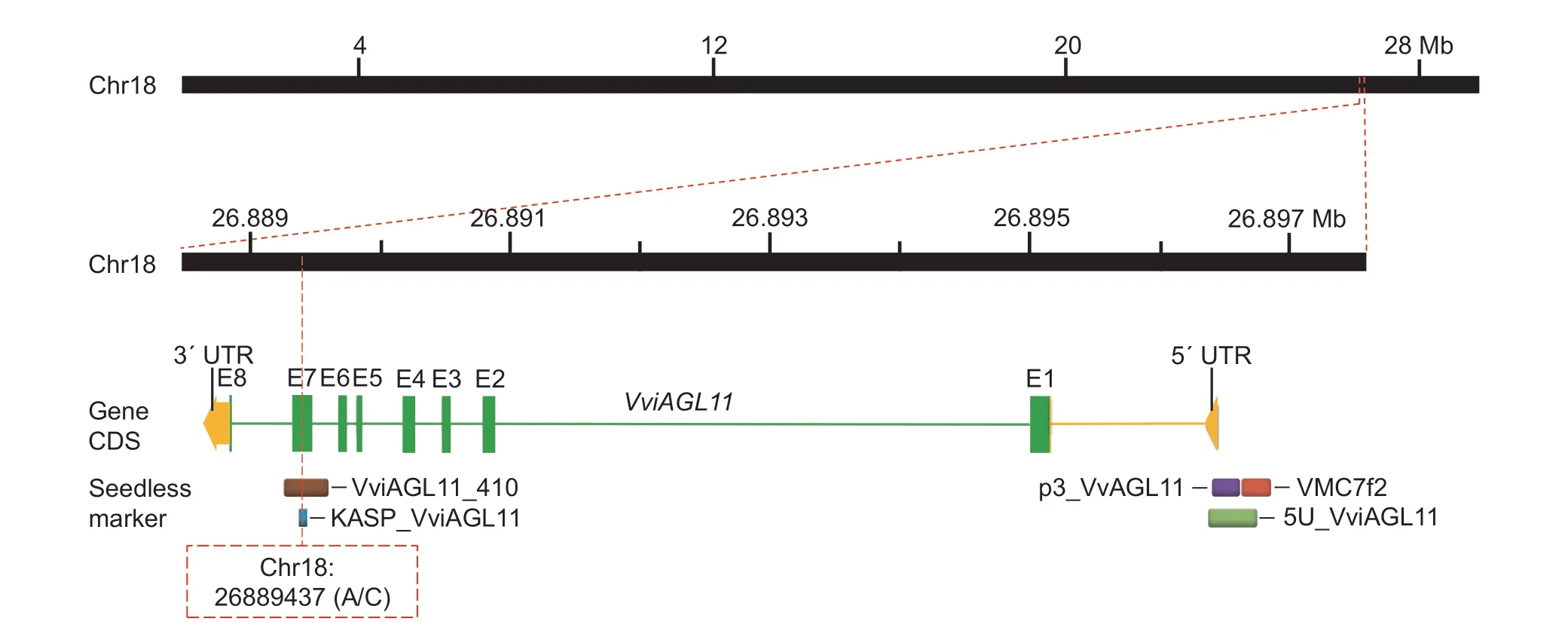
Fig.1 Physical locations of commonly used seedless markers on chromosome 18 of grapes (PN40024 12X).The main molecular markers providing high accuracy in identifying grapevines with seedless traits were VMC7f2 (red block), p3_VvAGL11 (purple block),5U_VviAGL11 (light green), KASP_VviAGL11 (blue block), and VviAGL11_410 (brown block).All of them were distributed in the VviAGL11 gene that was closely linked to the seedless trait of grape.The SSR markers VMC7f2, p3_VvAGL11 and 5U_VviAGL11 were located near the 5′ UTR (yellow block) of the VviAGL11 gene, with p3_VvAGL11 and 5U_VviAGL11 specifically located on the 5′ UTR of the VviAGL11 gene, which led to higher identification efficiency.The eight exons (E1–E8) of the VviAGL11 gene are shown in green blocks.The KASP_VviAGL11 molecular marker developed in this study was designed based on a mutation(A/C) of the base at Chr18: 26889437 located on E7 of the VviAGL11 gene, which was closely associated with the seedless trait in grape by Royo et al.(2018) through fine mapping.The amplified fragment of VviAGL11_410 was 410 bp, including the entire region of E7, which was used for Sanger sequencing to verify the reliability of KASP_VviAGL11.
On the other hand, single nucleotide polymorphism(SNP) markers offer significant advantages, including high-throughput detection, low cost and easy data interpretation.These benefits compensate for the technical limitations of SSR markers and have led to their increased attention and utilization by researchers(Wang et al.2022).Royo et al.(2018) found that the seedlessness phenotype is closely linked to a specific SNP located at Chr18: 26889437 (A/C) on exon 7 (E7)of the VviAGL11 gene, resulting in an Arg-197-Leu substitution (Fig.1).Subsequently, Ocarez et al.(2020)developed the e7_VviAGL11 marker based on this SNP,which efficiently and accurately identifies the seedless germplasm.However, the detection technology for this marker is quantitative PCR high resolution melting (qPCRHRM), which requires the use of the expensive StepOne Plus real-time PCR machine (Applied Biosystems,Foster City, CA, USA) and a special KAPA HRM FAST qPCR Master Mix enzyme (KAPABIOSYSTEMS,Cape Town, South Africa).An initial evaluation in this study revealed that the e7_VviAGL11 marker did not demonstrate sufficient cost advantages in detection compared to the p3_VvAGL11 marker.Therefore, after the consideration of many factors, we decided not to use the e7_VviAGL11 marker.Meanwhile, KASP (Kompetitive Allele Specific PCR), a detection technology based on the terminal bases of primers for specific matching of SNP genotyping and the detection of insertions and deletions(InDels), has become a commonly used type of SNP marker in MAS breeding due to its benefits of reduced time demand, labor and cost compared to qPCR-HRM detection technology (Semagn et al.2014; Grewal et al.2020).Wang et al.(2022) constructed an SNP fingerprint map of 348 grape cultivars using 46 high-quality KASP markers, which verified the feasibility of KASP technology for grape variety identification.To reduce breeding costs and improve the efficiency of the MAS breeding of seedless grape cultivars, it is useful to explore and design a KASP marker based on the SNP (A/C) at the Chr18: 26889437 position that was mapped by Royo et al.(2018).In addition, Sanger sequencing technology is considered to be the most direct and accurate method for detecting SNPs, with advantages such as strong targeting ability and accuracy of up to 100% (Wang et al.2020).Therefore, we designed a primer for the SNP(Chr18: 26889437 (A/C)) and used Sanger sequencing to determine the genotypes of different samples at this locus, which was necessary to verify the accuracy of the KASP markers and ensure the stability and reliability of the experimental results.
In the present study, we designed the KASP_VviAGL11 and VviAGL11_410 markers and compared them with the SSR markers p3_VvAGL11 and 5U_VviAGL11.We used these four markers to test 101 grape cultivars and 81 grape F1hybrid progenies.Then, we compared the results of different marker detections with the actual seed phenotypes and calculated indicators, such as the seedlessness detection rate and accuracy for different markers, to clarify the applicability of KASP_VviAGL11 in seedless grape MAS breeding.The results can provide more accurate and efficient molecular markers for seedless grape breeding, which is of great significance for simplifying and standardizing the MAS breeding process and accelerating the development of new seedless grape cultivars.
2.Materials and methods
2.1.Plant materials and DNA extraction
The experiment utilized leaves from 101 grape cultivars (48 seedless and 53 seeded) and 81 F1hybrid individual plants (49 seedless and 32 seeded) with different genetic backgrounds (Table 1).The grapesamples were obtained from the following sources:43 grape cultivars were collected in August 2021 from the Zhengzhou Grape Garden of National Fruit Tree Germplasm, Zhengzhou Institute of Fruit Research,Chinese Academy of Agricultural Sciences in Zhengzhou,Henan Province, China; 58 grape cultivars were obtained from the Grape Repository of Northwest A&F University in Yangling, Shaanxi Province, China (Table 1); 6 F1individual hybrid plants of ‘Flame Seedless’ (FS)בMuscat Hamburg’ (MH) were collected from Meixian Grape Test Station of Northwest A&F University in September 2022(Appendix A); 58 F1individual hybrid plants of ‘Red Globe’(RG)בDawn seedless’ (DS) were collected from Weishi Grape Test Station of Zhengzhou Fruit Research Institute,Chinese Academy of Agricultural Sciences in Kaifeng,Henan Province, China in September 2019 (Appendix B);and 17 F1individual hybrid plants of ‘SP522’ (SP)בFlame Seedless’ (FS) were collected in October 2021 from the Research Institute of Grape and Melon of Xinjiang Uygur Autonomous Region in Shanshan, Xinjiang Uygur Autonomous Region of China (Appendix C).
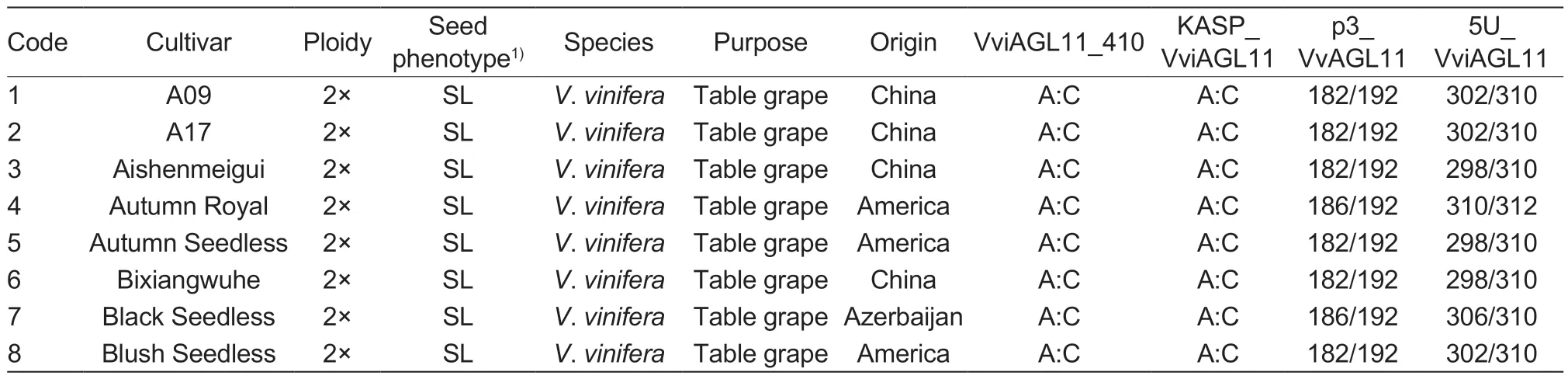
Table 1 Characteristics of the 101 grape (Vitis L.) cultivars used in this study
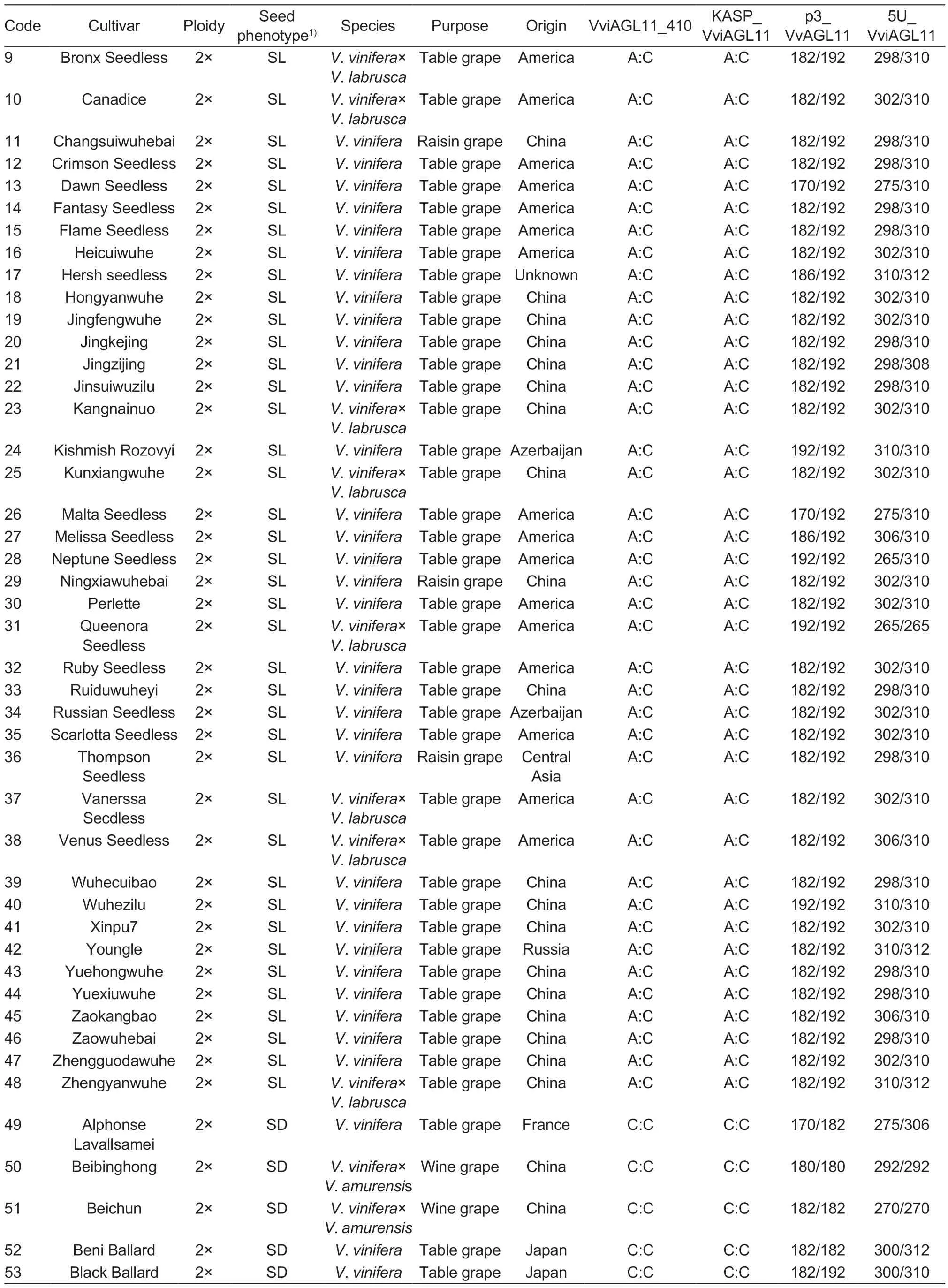
Table 1 (Continued from preceding page)
Genomic DNA was extracted from the leaves using a CTAB-based method (Li et al.2018).DNA concentration and purity were analyzed by agarose gel electrophoresis and spectrophotometry (NanoDrop 1000, Themo Scientific).The DNA was diluted to 50–100 ng μL–1and stored at –20°C.
2.2.Design and usage of the KASP_VviAGL11 marker
Firstly, we downloaded the DNA sequence containing 400 base pairs upstream and downstream of base pair 26 889 437 on chromosome 18 from the Grape Genome Browser (PN40024,12X) (https://www.genoscope.cns.fr/vitis/).Next, we designed the KASP_VviAGL11 marker primers using the Primer Premier 5.0 software to amplify a 67 bp fragment (Table 2).The forward primer a (Fa) is labeled with 5′-fluorescein-CE phosphoramidite (FAM)dye and specifically recognizes the A allele, while the forward primer b (Fb) primer is labeled with 5′-hexachlorofluorescein-CE phosphoramidite (HEX) dye and specifically recognizes the C allele.The KASP primers were resuspended in 10 mmol L–1TE (pH 8.0) to 50 μmol L–1.The forward primers Fa and Fb and the reverse universal primer R were mixed in a ratio of 1:1:3 to prepare the KASP Primer Mix.The KASP-PCR reaction system comprised 2 μL template DNA, 0.125 μL KASP Primer Mix, and 5 μL 2× KASP Master Mix, supplemented with nuclease-free water to a final volume of 10 μL (Yang et al.2021).The KASP Master Mix was FLU-ARMS for KASP 2× PCR mix (STO ROX).The KASP-PCR reactions included the following steps: (1) pre-denaturation at 94°C for 15 min; (2) 10 cycles of denaturation at 94°C for 20 s,followed by annealing at 61–55°C for 60 s, with each cycle reduced by 0.6°C; (3) 26 cycles of denaturation at 94°C for 20 s, followed by annealing and extension at 55°C for 60 s; and (4) fluorescence reading at 37°C for 1 min.The first three stages were performed on an ABI 9800 FAST Thermocycler, and fluorescence was read with an ABI 7900 Real-Time PCR System (Applied Biosystems).All the reagents and equipment used above were obtained from Guangzhou Goodall Biotechnology Co., Ltd.
2.3.Validation of the accuracy of the KASP_VviAGL11 marker
In order to validate the accuracy of the developed KASP_VviAGL11 marker, the VviAGL11_410 marker was designed using Primer Premier 5.0.The 410 bp DNA sequence located from Chr18: 26889269–26889678 was amplified using conventional PCR technology, followed by Sanger sequencing analysis to confirm that the genotype of the sample DNA at position Chr18: 26889437 aligned with the genotype detected by the KASP_VviAGL11 marker.The primer sequences for VviAGL11_410 were Forward 5′-GGCTACTTGGTGATTTATGTGCTCT-3′ and Reverse 5′-GAAGGCACAACAGTTGATACCGATC-3′.The conventional PCR reaction system had a total volume of 50 μL, including 2 μL template DNA at 50 ng μL–1, 1 μL of each forward and reverse primer (10 μmol L–1), 1 μL TransStart?FastPfu DNA Polymerase (2.5 units μL–1),10 μL 5× TransStart?FastPfu Buffer, 4 μL dNTP mixture(2.5 mmol L–1), and 31 μL nuclease-free water (Li et al.2021).The PCR reaction program consisted of an initial denaturation step at 95°C for 2 min, followed by 32 cycles of denaturation at 95°C for 20 s, annealing at 58°C for 20 s, extension at 72°C for 30 s, and a final elongation at 72°C for 5 min.The primer design and sequencing were carried out by Sangon Biotech (Shanghai) Co., Ltd.

Table 2 Primer sequences for KASP_VviAGL11
2.4.Amplification and detection of SSR markers p3_VvAGL11 and 5U_VviAGL11
In order to determine whether KASP_VviAGL11 was superior to other grape seedless markers, we performed DNA detection on all samples using the SSR markers p3_VvAGL11 and 5U_VviAGL11, which are recognized for their high accuracy in identifying grape seedlessness.The primer sequences for p3_VvAGL11 were Forward 5′-CTCCCTTTCCCTCTCCCTCT-3′ and Reverse 5′-AAACGCGTATCCCAATGAAG-3′ (Mejía et al.2011).Similarly, the primer sequences for 5U_VviAGL11 were Forward 5′-CGCCCATTCTCTCTCGCTAT-3′ and Reverse 5′-GTGCAAAAACGCGTATCCCA-3′ (Ocarez et al.2020).The forward primers for p3_VvAGL11 and 5U_VviAGL11 were labeled with FAM and HEX fluorescent probes,respectively.The PCR reaction system for the SSR markers had a total volume of 10 μL, consisting of 5 μL 2× Taq Master Mix for PAGE, 0.5 μL of each forward and reverse primer (10 μmol L–1), 2 μL DNA template (10 ng μL–1), and 2 μL nuclease-free water (Chen et al.2021).The reaction program included an initial denaturation step at 94°C for 3 min, followed by 29 cycles of denaturation at 94°C for 30 s, annealing at 59°C for 30 s, extension at 72°C for 40 s, and a final extension at 72°C for 5 min.The amplified SSR marker products were analyzed through the 3730XL genetic analyzer, and GeneMapper 4.0 software was used for data analysis.The synthesis and detection of SSR primers were completed by Beijing Novogene Science and Technology Co., Ltd.
2.5.Data analysis
According to the seed characteristics, grapes can be classified into two categories for statistical analysis:seedless (including parthenocarpy and stenospermocarpy types) and seeded (representing fully developed grape seeds) (Pratt 1971; Ledbetter and Ramming 1989;Bouquet and Danglot 1996).Different markers were tested to evaluate true positive (TP), true negative (TN),false positive (FP), and false negative (FN) indicators(Vihinen 2012; Bennici et al.2019).FP refers to the detection of seedless alleles in a seeded sample,while FN indicates the absence of seedless alleles in a seedless sample.We calculated the false positive rate (FPR)=FP/(TP+FP+TN+FN), false negative rate (FNR)=FN/(TP+FP+TN+FN), accuracy rate(AR)=(TP+TN)/(TP+FP+TN+FN), and seedless detection rate (SDR)=TP/(TP+FP).To evaluate the association between phenotype and detection data, a chi-square(χ2) test for independence (P≤0.01) was performed using SPSS 26.0 software to analyze the relationship between the phenotype and the allele or genotype of the different markers (Chen et al.2021).
3.Results
3.1.Validation of the universality of the KASP_VviAGL11 marker on 101 grape cultivars
In this study, the KASP_VviAGL11 marker was used for the genotyping of 101 collected grape cultivars (Table 1).The analysis of the grape cultivars detected two alleles, A and C, with the A allele (χ2=101.00, P=9.198E–24) being significantly associated with the seedless phenotype of grapes (Table 3).All 48 seedless grape cultivars featured the A allele, and their corresponding genotype was A:C(χ2=101.00, P=9.198E–24) (Table 4).In contrast, all 53 seeded grape cultivars had the C:C genotype (χ2=101.00,P=9.198E–24).It is noteworthy that the A allele was detected in all seedless grape cultivars, indicating a 100%detection and accuracy rate (Table 5).
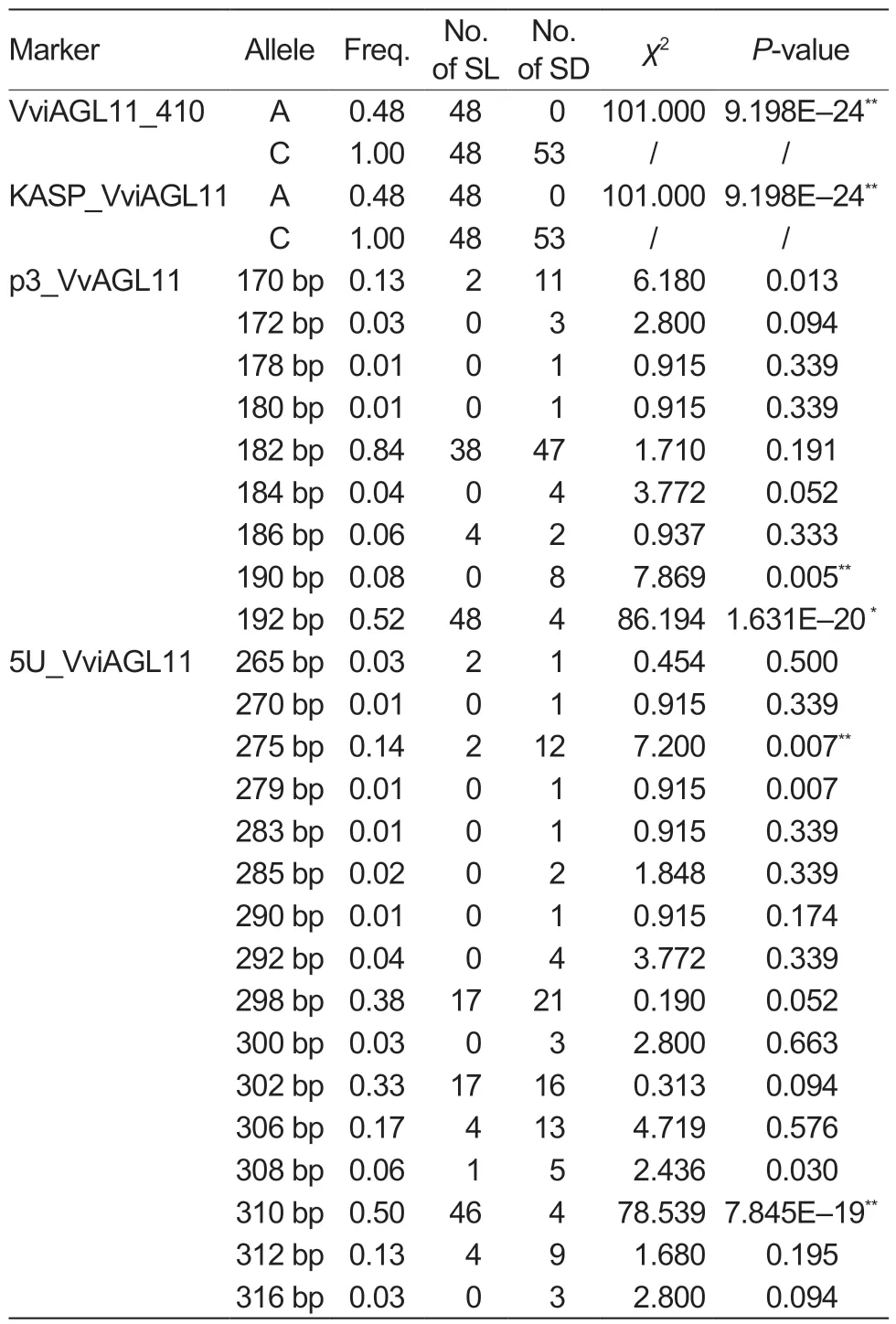
Table 3 Correlation analysis between the distribution of allele types of the different markers and the seedless trait in 101 grape cultivars1)
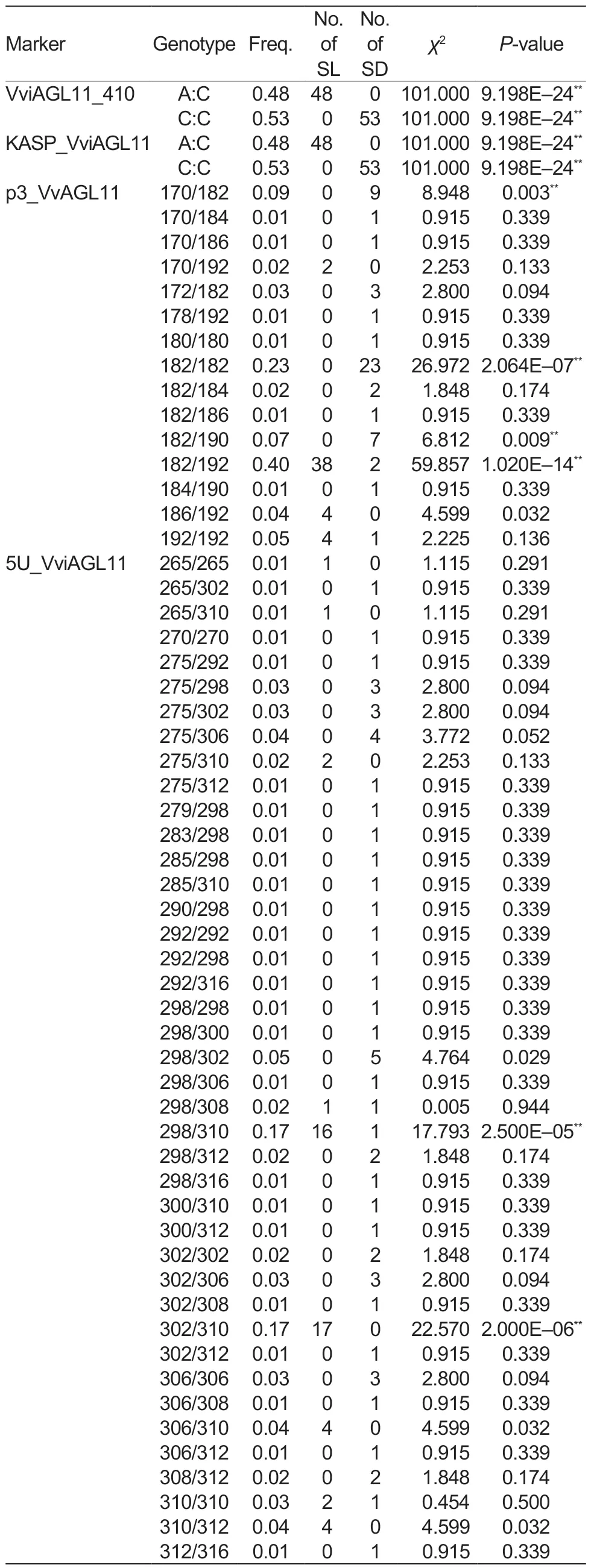
Table 4 Correlation analysis between the distribution of genotypes with different markers and the seedless trait in 101 grape cultivars1)
In order to validate the reliability of the KASP_VviAGL11 marker, we developed the VviAGL11_410 marker and tested it on 101 different grape cultivars.We conducted Sanger sequencing and found that at the position Chr18:26889437, the 48 seedless grape cultivars all possessed the A:C genotype (χ2=101.00, P=9.198E–24) (Table 4),while the genotypes of 53 seeded grape cultivars were all C:C (χ2=101.00, P=9.198E–24), which was fully consistent with the detection results of the KASP_VviAGL11 marker.Therefore, it is reasonable to conclude that if the A allele is detected using the KASP_VviAGL11 marker in grapes,it can be identified as a seedless trait.
3.2.Evaluation of the detection efficiencies of p3_VvAGL11 and 5U_VviAGL11 across 101 grape cultivars
Testing of the 101 grape cultivars with the SSR molecular marker p3_VvAGL11 revealed a total of nine alleles, with sizes of 170, 172, 178, 180, 182, 184, 186, 190, and 192 bp (Table 3).Among them, the 190 bp (χ2=7.869,P=0.005) and 192 bp (χ2=86.194, P=1.631E–20) alleles showed strong associations with the seed phenotype.The 192 bp allele was only found in 48 seedless grape cultivars and four seeded grape cultivars, indicating that it was the allele most significantly associated with seedlessness.Among the 15 genotypes detected by the p3_VvAGL11 marker, the 182/192 genotype(χ2=59.857, P=1.020E–14) was significantly associated with the seedless phenotype, since it was detected in 38 seedless cultivars.The remaining seedless cultivars expressed the 170/192 (χ2=2.253, P=0.133), 186/192(χ2=4.599, P=0.032), and 192/192 (χ2=2.225, P=0.136)genotypes (Table 4).In other words, the 192 bp allele was detected in all seedless grape cultivars, with a detection rate of 92.31% and an accuracy rate of 96.04%(Table 5).These findings suggested that the presence of the 192 bp allele detected by the p3_VvAGL11 marker could reliably identify the seedless trait in the grape samples.
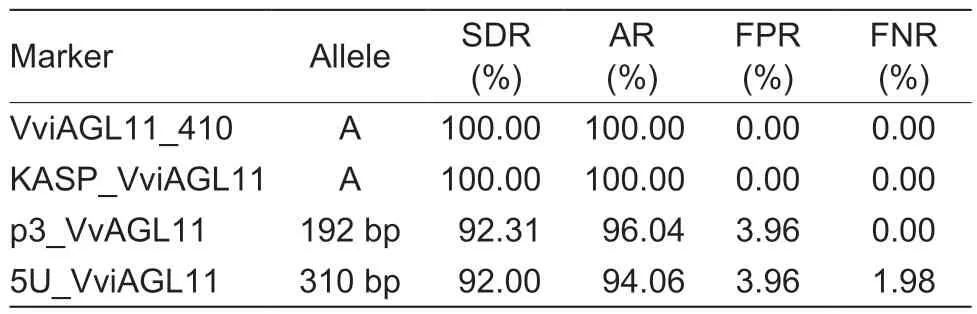
Table 5 Identification efficiency of alleles with different markers that are closely associated with the seedless trait in 101 grape cultivars1)
The SSR marker 5U_VviAGL11 detected 16 alleles in all 101 grape cultivars, with sizes of 265, 270, 275,279, 283, 285, 290, 292, 298, 300, 302, 306, 308,310, 312, and 316 bp (Table 3).Among them, the 275 bp (χ2=7.200, P=0.007) and 310 bp (χ2=78.539,P=7.845E–19) alleles showed highly significant associations with the seed phenotype.The 310 bp allele was only detected in 46 seedless grape cultivars and four seeded grape cultivars, while it was absent from the remaining 49 seeded cultivars.This strongly suggested that the 310 bp allele is closely associated with the seedless phenotype.Among the 41 genotypes identified by the 5U_VviAGL11 marker, the 298/310(χ2=17.793, P=2.500E–05) and 302/310 (χ2=22.570,P=2.000E–06) genotypes were detected in 33 seedless cultivars, indicating their significant link to the seedless phenotype.The remaining 15 seedless cultivars expressed the 265/265 (χ2=1.115, P=0.291), 265/310(χ2=1.115, P=0.291), 275/310 (χ2=2.253, P=0.133),298/308 (χ2=0.005, P=0.944), 306/310 (χ2=4.599,P=0.032), 310/310 (χ2=0.454, P=0.500), and 310/312(χ2=4.599, P=0.032) genotypes (Table 4).From the above data, the 310 bp allele was detected in 95.83%of the seedless cultivars, resulting in a detection rate of 92.00% and an accuracy rate of 94.06% (Table 5).Therefore, it is reasonable to conclude that when the 310 bp allele is detected by the 5U_VviAGL11 marker,the grape variety can be accurately identified as seedless.
3.3.Comparative analysis of the KASP_VviAGL11 marker using the VviAGL11_410, p3_VvAGL11, and 5U_VviAGL11 markers in 101 grape cultivars
After the analyses in Sections 3.1 and 3.2, we identified the specific alleles associated with the seedless trait in grapes using the KASP_VviAGL11, VviAGL11_410, p3_VvAGL11, and 5U_VviAGL11 markers, which were A, A,192 bp, and 310 bp, respectively.Then, we assessed the efficiency of each marker in detecting these specific alleles.The results revealed that both the KASP_VviAGL11 and VviAGL11_410 markers exhibited 100%detection and accuracy rates with no false positives or negatives, outperforming the p3_VvAGL11 marker that showed a false positive rate of 3.96%.In contrast, the 5U_VviAGL11 marker produced not only a false positive rate of 3.96% but also a false negative rate of 1.98%(Table 5).Therefore, we concluded that the KASP_VviAGL11 marker offers higher accuracy compared to the p3_VvAGL11 and 5U_VviAGL11 markers, and its validation using the VviAGL11_410 marker enhances its stability and reliability.
3.4.Validation of KASP_VviAGL11, VviAGL11_410,p3_VvAGL11, and 5U_VviAGL11 in the grape F1 hybrid population
In order to further validate the identification ability of the KASP_VviAGL11 marker across different genetic backgrounds, we selected a total of 81 individuals from three grape F1hybrid populations.One population was obtained through embryo rescue technology from the cross FS×MH, while the other two were conventional hybrid breeding populations resulting from the crosses RG×DS and SP×FS.All 81 individuals could be detected by the KASP_VviAGL11, VviAGL11_410, p3_VvAGL11,and 5U_VviAGL11 markers (Fig.2).
After testing six hybrid F1offspring of FS×MH (three seedless and three seeded individuals), we found that KASP_VviAGL11 and VviAGL11_410 were able to detect three genotypes (A:A, A:C, and C:C).The A allele was only present in the three seedless individuals (χ2=6.000,P=0.014), while none of the three seeded individuals had the A allele.As a result, the seedless detection rate and accuracy for the six individuals were both 100%, with 0%false positives or false negatives.Using p3_VvAGL11,we identified two genotypes (182/192 and 192/192)and found the 192 bp allele in all individuals (χ2=0.000,P=1.000), yielding a seedless detection rate and accuracy of 50%, along with a 50% false positive rate.With the same efficiency as the p3_VvAGL11 marker,using 5U_VviAGL11 allowed us to identify two genotypes(298/310 and 310/310), with the 310 bp allele detected in all individuals (χ2=0.000, P=1.000) (Table 6; Appendix A).
After testing 58 hybrid F1offspring of RG×DS (39 seedless and 19 seeded individuals), we found that the KASP_VviAGL11 and VviAGL11_410 markers were able to detect two genotypes (A:C and C:C).Specifically,only the 39 seedless individuals contained the A allele(χ2=58.000, P=2.621E–14), while none of the 19 seeded individuals possessed it.Consequently, the seedless detection rate and accuracy both achieved 100%,with 0% false positives or false negatives.Using p3_VvAGL11, we identified two genotypes (170/182 and 182/192), with the 192 bp allele identified in 37 seedless individuals (χ2=49.785, P=1.715E–12), while the 192 bp allele was not detected in the remaining individuals.As a result, the 192 bp allele was detected in 94.87% of the seedless individuals, leading to a seedless detection rate of 100% and an accuracy of 96.55%, with 3.45% false negatives.Similarly, using 5U_VviAGL11 resulted in the identification of four genotypes (275/298, 275/302,298/310, and 302/310), with the 310 bp allele detected in 37 seedless individuals (χ2=49.785, P=1.715E–12),while the 310 bp allele was not found in the remainingindividuals.Therefore, the detection efficiency was consistent with that of the p3_VvAGL11 marker (Table 6;Appendix B).
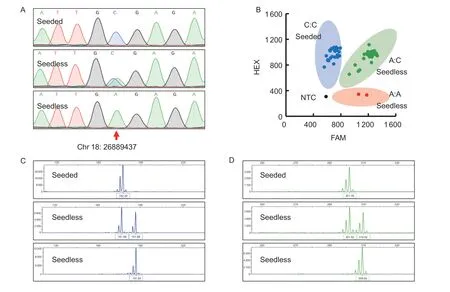
Fig.2 Examples of different markers used for identifying the seedless trait in grapevines.A, the genotype at position Chr18:26889437 on the grapevine chromosome was determined by amplifying the VviAGL11_410 marker using PCR, followed by Sanger sequencing and sequence alignment, where C:C represents the seeded phenotype and A:C or A:A represent the seedless phenotype.B, the KASP_VviAGL11 marker was used for amplification from 81 F1 hybrids of different genetic backgrounds, and the fluorescence values of different samples were read on an ABI 7900 fluorescence detector to determine the genotypes.The signal points near the vertical axis indicate the detection of a strong HEX signal, which corresponds to the genotype C:C (blue dot) and indicates a seeded phenotype.The signal points near the horizontal axis indicate that the FAM fluorescence signal was strong,and the corresponding genotype was A:A (red dot), indicating a seedless phenotype.The signal points in the middle indicate the detection of both strong FAM and HEX signals, corresponding to the genotype A:C (green dot), which also means that they were seedless.The black dot represents a negative control (no DNA samples added), with weak FAM and HEX signals indicating no alleles.C, the p3_VvAGL11 marker was used for DNA amplification, followed by the analysis of fluorescent fragment patterns using the ABI3730XL automated genetic analyzer and GeneMapper v3.25 software.The representative fragment 182/182 indicates a seeded phenotype, while 182/192 or 192/192 indicate a seedless phenotype.D, the 5U_VviAGL11 marker was used for DNA amplification, followed by the same analysis as in C.The representative fragment 302/302 indicates a seeded phenotype, while 302/310 or 310/310 indicate a seedless phenotype.
After testing 17 hybrid F1progenies of SP×FS (seven seedless and 10 seeded individuals), we found that both the KASP_VviAGL11 and VviAGL11_410 markers were able to detect two genotypes (A:C and C:C).Only the seven seedless individuals carried the A allele (χ2=17.000,P=3.700E–05), while none of the 10 seeded individuals had it.Consequently, the seedless detection rate and accuracy were both 100%, with 0% false positives or false negatives.Using p3_VvAGL11, we identified five genotypes (182/182, 182/186, 182/192, 186/186, and 186/192), with the 192 bp allele detected in only the seven seedless individuals (χ2=17.000, P=3.700E–05).Similarly, using 5U_VviAGL11 resulted in the identification of seven genotypes (298/302, 298/306, 302/302,302/306, 302/310, 306/306, and 306/310), with only the seven seedless individuals presenting the 310 bp allele(χ2=17.000, P=3.700E–05).Therefore, the detection efficiencies of both the p3_VvAGL11 and 5U_VviAGL11 markers were consistent with that of the KASP_VviAGL11 marker (Table 6; Appendix C).
In summary, we have demonstrated that the KASP_VviAGL11 and VviAGL11_410 markers exhibit high accuracy and stability across different genetic backgrounds, while the p3_VvAGL11 and 5U_VviAGL11 markers show some degree of difference in efficiency.
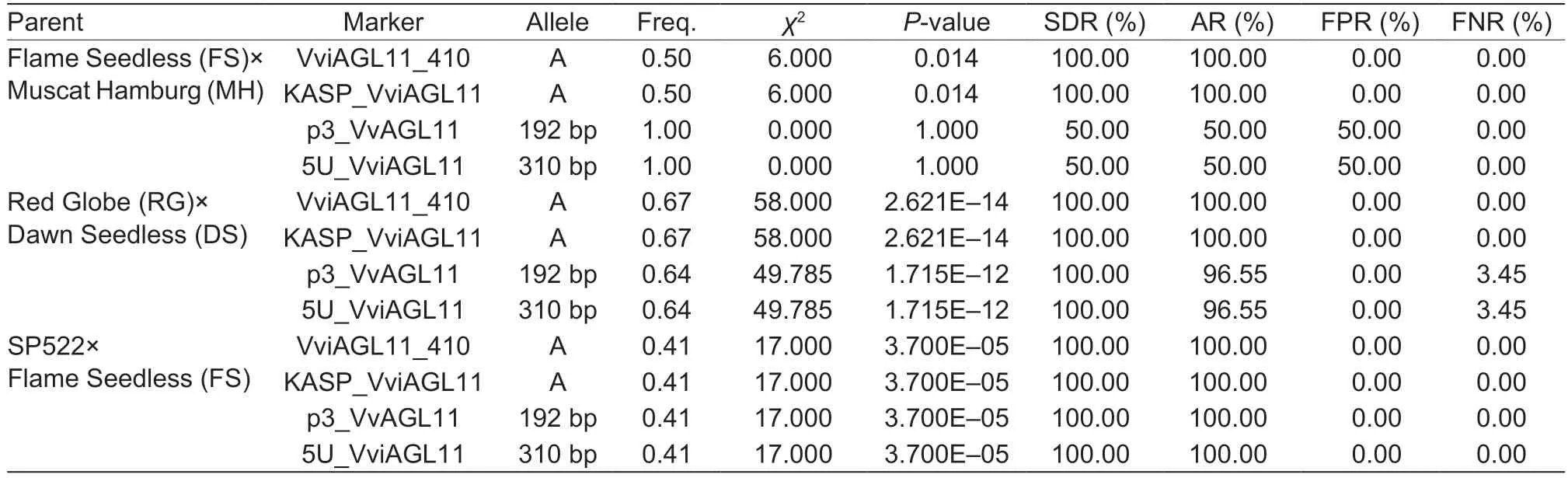
Table 6 Analysis of identification efficiency of the different molecular markers for F1 hybrids of different combinations1)
3.5.Comparative analysis of the KASP, Sanger sequencing and SSR detection technologies
In order to identify the most cost-effective, efficient and accurate marker, we conducted a comparative analysis of the KASP, Sanger sequencing and SSR detection technologies used for the KASP_VviAGL11,VviAGL11_410 and p3_VvAGL11 markers, respectively,considering both experimental time and cost (Fig.3;Appendix D).In terms of cost, the KASP detection technology is the most economical of the three, followed by SSR detection, while Sanger sequencing is the most expensive technology.With respect to detection efficiency, KASP is more than twice as efficient compared to the other two technologies.In terms of accuracy, both KASP and Sanger sequencing can achieve 100%, while SSR detection has slightly lower accuracy due to its potential for false positives or negatives.In terms of data analysis, KASP detection technology can directly generate genotypes, making the data analysis process easier.In contrast, the results obtained from Sanger sequencing require the confirmation of sample genotypes through blast analysis, making data analysis more complex and time-consuming.Additionally, SSR detection technology is susceptible to errors, which can make data comparison difficult.In terms of the visualization of detection results,KASP can directly display the genotypes of all samples in a clustering diagram, which is especially useful for analyzing trait classification in hybrid populations.It has the best display effect among the three detection technologies.In contrast, the Sanger sequencing and SSR detection technologies require checking the detection result of each sample individually before summarizing and displaying the data, resulting in a lower visualization ability.In conclusion, we believe that using the KASP detection technology described in this study offers greater advantages.
4.Discussion
Seedless grape cultivars can be classified into two types based on their pollination and fruiting characteristics:parthenocarpic and pseudo-parthenocarpic (also known as stenospermacarpic) (Pratt 1971; Ledbetter and Ramming 1989).The ovules of parthenocarpic grapes directly develop into fruit without fertilization, resulting in smaller fruit without seeds.In contrast, stenospermacarpic grapes have fertilized embryos that abort during the later stages of fruit development, leaving seed residues in the berry and yielding seedless fruit.Stenospermacarpic grapes are generally larger than parthenocarpic grapes and are commonly used in hybrid breeding and cultivation, such as the ‘Thompson Seedless’, ‘Ruby Seedless’, ‘Flame Seedless’, and ‘Crimson Seedless’cultivars (Akkurt et al.2019).Both parthenocarpic and pseudo-parthenocarpic grapes eliminate the concern of hard seeds during consumption, with little difference in texture.Furthermore, researchers commonly classify grape seed traits as seeded or seedless using binary processing statistics during the breeding process(Bergamini et al.2013; Ocarez et al.2020).Therefore,our study also collectively refers to parthenocarpic and pseudo-parthenocarpic grapes as seedless grapes, which are classified into seeded or seedless grapes based on the seed phenotype rather than using the seedless level classification method of Bennici et al.(2019).This choice ensures that the research results are more closely related to the actual breeding goals.
There are two main approaches for seedless grape breeding: conventional hybrid breeding and the embryo rescue technique (Fig.4).The former uses a seeded grape cultivar as the female parent and a seedless grape cultivar as the male parent, but the probability of obtaining seedless offspring is very low (Bouquet and Danglot 1996).The embryo rescue techniqueuses seedless material as the female parent and cultures the embryos from pollination in vitro to obtain hybrid offspring.This method has a higher probability of obtaining seedless offspring but features a lower success rate (Li et al.2015).Both methods for breeding seedless grapes are time-consuming and laborintensive.The breeding process can be expedited with the use of accurate and efficient molecular markers in the early selection of hybrid offspring, which can also reduce management costs.Previous research has revealed that the SSR markers p3_VvAGL11 and 5U_VviAGL11 have a higher frequency and accuracy in identifying seedlessness (Chen et al.2021).This study involving 101 grape cultivars confirmed that the SSR markers p3_VvAGL11 and 5U_VviAGL11 are closely associated with the seedless trait when detecting alleles 192 bp (χ2=86.194, P=1.631E–20) and 310 bp(χ2=78.539, P=7.845E–19).These findings differ from previous studies regarding which alleles are closely linked to seedlessness, indicating that SSR markers may be susceptible to data errors and challenging to use for analysis and comparison with other researchers’ results.
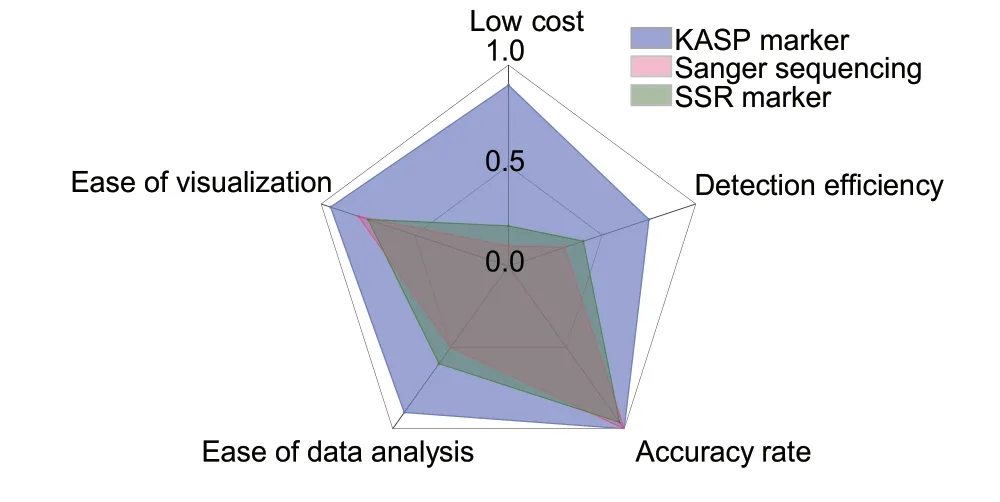
Fig.3 Comparative analysis of the main characteristics of the three detection techniques.The data in this figure were sourced from the B values in Appendix D.
Both the p3_VvAGL11 and 5U_VviAGL11 markers exhibited false positives in four seeded cultivars,namely, ‘Black Ballard’, ‘Golden Finger’, ‘Moldova’, and‘Zaomeigui’.In addition, the 5U_VviAGL11 marker produced two false negatives in seedless cultivars,namely, ‘Jingzijing’ and ‘Queenora Seedless’, which were consistent with the findings reported by Chen et al.(2021).These results demonstrate that the detection of p3_VvAGL11 and 5U_VviAGL11 markers is prone to false positives, while they also confirm the reliability of the data obtained in this work.
In the present study, we developed the KASP_VviAGL11 and VviAGL11_410 markers to address the limitations of SSR markers and improve the accuracy of seedless grape detection.The results confirmed that both markers are closely linked to the seedless trait when detecting the A allele, with VviAGL11_410 serving as a validation marker and achieving 100% accuracy for KASP_VviAGL11.Through a comparison of the locations of all four markers in the VviAGL11 gene, it can be inferred that the KASP_VviAGL11 and VviAGL11_410 markers were designed based on the single nucleotide mutation (Chr18: 26889437 (A/C)) of Arg-197-Leu in the VviAGL11 gene, which is considered to be the most critical site for forming seedlessness, while the p3_VvAGL11 and 5U_VviAGL11 markers are located on the 5′ UTR of the VviAGL11 gene (Fig.1).Previous studies have suggested that the main reason for false positives is the inability of the VviAGL11 gene to express the seedless trait, even though the 5′ UTR carries the seedless mutation sequence in the absence of the Arg-197-Leu mutation in the coding region (Amato et al.2022).On the other hand, false negatives are caused by other minor genes responsible for regulating the seedless trait (Ocarez et al.2020), which ultimately result in differences in accuracy between the markers.We subsequently compared the KASP, Sanger sequencing and SSR detection techniques used in this study from the following five perspectives: cost-effectiveness,detection throughput, accuracy, difficulty of data analysis,and visualization effects.We concluded that the KASP technology is simpler and more economical, efficient,and precise, which is consistent with the research findings of Semagn et al.(2014) and Zhao et al.(2017).It is worth noting that the grape samples used in this study included common cultivars from different genetic backgrounds to make the experimental results more robust and applicable.For instance, the table grapes‘Ruby Seedless’ and ‘Shine Muscat’ belong to the species V.vinifera and V.vinifera×V.labrusca, respectively.Similarly, the wine grapes ‘Pinot Noir’ and ‘Beibinghong’are classified as V.vinifera and V.vinifera×V.amurensis,respectively.Therefore, we strongly recommend giving priority to the use of the KASP_VviAGL11 marker to identify the seedless trait in grapes.
This study unexpectedly detected two homozygous A:A genotypes (‘8_1_3’ and ‘8_10_2’) during the examination of the FS×MH hybrid F1generations using KASP_VviAGL11 (Fig.2-B).According to genetic laws,it is impossible for a homozygous A:A genotype to be obtained from the hybridization of maternal FS (A:C) and paternal MH (C:C).However, this phenomenon may have occurred due to self-pollination of the maternal FS(A:C).Although the homozygous A:A genotype seedless material is particularly challenging to obtain, it holds significant value as an important material for seedless grape breeding.For instance, crossbreeding with homozygous A:A genotype seedless material can greatly increase the rate of seedless offspring.Additionally, inthe previous stage of this study, polyploid seedless materials were utilized to verify four markers, but none of them were successful in detecting the seedless allele(Appendix E).This result reveals that none of the studied markers, including KASP_VviAGL11, VviAGL11_410, p3_VvAGL11,and 5U_VviAGL11, are appropriate for identifying polyploid seedless grapes, and further suggests that the seedless trait in grapes is a complex characteristic that is influenced by multiple genes.
Overall, our study has successfully optimized the process of MAS breeding for diploid seedless grapes, ensuring a 100% selection rate for seedless hybrid individuals (Fig.4).The optimization process involves the following five steps:(1) culturing hybrid seedlings obtained through conventional sexual hybridization or embryo rescue technology to the stage of 5–7 true leaves; (2) selecting the fourth true leaf of the young seedling, grinding it in a 2.0 mL centrifuge tube and extracting DNA; (3) amplifying the DNA of different seedlings using the KASP_VviAGL11 marker and identifying the seedless germplasms with A:A (red dot) or A:C(green dot) genotypes, while those with C:C (blue dot) genotypes are identified as seeded germplasms; (4) conducting spot checks on the seedless germplasms identified by the KASP_VviAGL11 marker using VviAGL11_410 to ensure 100%accuracy of the screening results; and (5)transplanting the finally selected seedless germplasm to the field for management.The above process offers various advantages, including simplicity, efficiency and low cost, which compensate for the shortcomings of using SSR seedless molecular markers for MAS breeding and significantly enhance the efficiency of seedless breeding.
5.Conclusion
The KASP_VviAGL11 marker developed in this study can detect diploid seedless grape germplasms with 100% accuracy,which optimizes the MAS breeding process for seedless grapes, while iteffectively reduces the cost of seedless grape breeding and significantly improves its efficiency.
Acknowledgements
This work was supported by the earmarked fund for the China Agriculture Research System of MOF and MARA (CARS-29-yc-3), the Project of Agricultural Breeding in Ningxia Hui Autonomous Region,China (NXNYYZ20210104) and the Key Industrial Innovation Chain Project in Shaanxi Province, China(2021ZDLNY04-08).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.10.014
 Journal of Integrative Agriculture2023年11期
Journal of Integrative Agriculture2023年11期
- Journal of Integrative Agriculture的其它文章
- Germplasm and molecular breeding in horticultural crops
- QTL analysis of early flowering of female flowers in zucchini(Cucurbita pepo L.)
- A novel mutation in ACS11 leads to androecy in cucumber
- Comprehensive analysis of the full-length transcripts and alternative splicing involved in clubroot resistance in Chinese cabbage
- Virucidal activity of MICRO-CHEM PLUS against African swine fever virus
- Effects of the combined application of organic and chemical nitrogen fertilizer on soil aggregate carbon and nitrogen: A 30-year study
