Multiparametric Imaging and Nanomechanical Analysis of Single Native Virus Particles Under Aqueous Conditions by Atomic Force Microscopy*
YANG Yan-Qi, FENG Ya-Qi, WEI Jia-Jia, LI Mi**
(1)State Key Laboratory of Robotics, Shenyang Institute of Automation, Chinese Academy of Sciences, Shenyang 110016, China;2)Institutes for Robotics and Intelligent Manufacturing, Chinese Academy of Sciences, Shenyang 110169, China;3)University of Chinese Academy of Sciences, Beijing 100049, China)
Abstract Objective Detecting the detailed behaviors of single viruses is essential for uncovering the underlying mechanisms guiding virus life cycle, which significantly benefits developing therapeutic methods against viral infection. The advent of atomic force microscopy (AFM) provides a novel powerful tool to characterize the structures and mechanical properties of single viruses with unprecedented spatial resolution, and applications of AFM in single-virus assay have contributed much to the field of physical virology. Nevertheless, the mechanical cues of single native viruses during viral activities are still not fully understood, and particularly studies of utilizing multiparametric AFM imaging to investigate the behaviors of single viruses are still scarce. Here,multiparametric AFM imaging was combined with AFM indentation assay to investigate the structural and mechanical dynamics of single native virus particles in response to chemical stimuli under aqueous conditions. Methods The poly-L-lysine was used to coat the coverslips to attach lentivirus particles onto the coverslips, and then the virus particles were probed by AFM in pure water. Single virus particles were imaged at the peak force tapping (PFT)-based multiparametric AFM imaging mode, in which the topographical images and mechanical maps of the virus particles were obtained simultaneously. Under the guidance of AFM’s topographical imaging, the AFM probe was moved to the central area of the virus particle to perform indentation assay for measuring the mechanical properties of the virus. The alcohol solution (75%) was used as an example of chemical stimulus to treat virus particles,after which the structural and mechanical changes of individual virus particles were revealed by AFM. Results The structures and mechanical properties of single virus particles could be well characterized by AFM under aqueous conditions, and the virus particles exhibited different elastic and adhesive properties in air and in liquid. After the treatment of alcohol, the shape of virus particles became irregular, and the virus particles became stiffer as well as less deformable. Conclusion The research provides a novel way to investigate the structures and nanomechanical properties of single native viruses in liquids based on AFM, which will have general implications for the field of virology.
Key words atomic force microscopy (AFM), virus particle, multiparametric AFM imaging, nanomechanical properties, force curve, Young’s modulus
It is well known that viruses play an important role in the life activities and pathological processes of biological systems. Horizontal gene transfer (HGT),which is the sharing of genetic material between organisms that are not in a parent-offspring relationship, has long been recognized as an important force in the evolution and biodiversity of global ecosystems[1]. Bacteria acquire novel DNA through HGT, which enables them to adapt to changing environmental conditions, and viruses are an important mediator for bacterial HGT[2]. The virusinfected cell is often killed by the massive proliferation of virus particles inside it, but sometimes, the viral DNA, instead of directly generating virus particles, may persist in its host for many cell generations as a relatively innocuous passenger (either as a separate intracellular fragment of DNA, known as a plasmid, or as a sequence inserted into the cell’s regular genome) to contribute the transfers of genetic materials between organisms[3]. A recent study by Irwinet al.[4]systematically characterized viral-eukaryotic gene exchange and identified thousands of transfers which were associated with novel functionality, evidencing that viruses widely participate in eukaryotic evolution.Despite the great contributions of viruses in HGT for biodiversity, an undisputable fact is that some viruses are detrimental or even deadly to human beings. Over the past years, several notorious viruses have been causing extreme panic and heavy burden to human life in the world, including Zika virus[5], Ebola virus[6],severe acute respiratory syndrome coronavirus 2(SARS-CoV-2)[7], and so on. Viruses are also associated with human cancers, and so far, seven human viruses have been found to cause 10%-15% of human cancers worldwide, including Epstein-Barr virus (EBV) for Burkitt’s lymphoma, hepatitis B virus (HBV) for hepatocellular carcinoma, human T-lymphotropic virus-I (HTLV-I) for adult T cell leukemia, human papillomavirus (HPV) for cervical cancer and penile cancer, hepatitis C virus (HCV) for hepatocellular carcinoma, Kaposi’s sarcoma herpesvirus (KSHV) for Kaposi’s sarcoma, and Merkel cell polyomavirus (MCV) for Merkel cell carcinoma[8]. Consequently, investigating the underlying mechanisms guiding the activities of viruses is of fundamental significance for treating virus-induced diseases.
The advent of atomic force microscopy (AFM)provides a powerful tool to characterize the structural and mechanical behaviors of single native viruses. In recent years, it is increasingly apparent in the communities of virology that detecting the behaviors of single viruses benefits probing specific virus groups and revealing genomic microdiversity within viral populations[9]. Single-virus assay has become an important strategy for the investigations of viral activities, which significantly complements traditional ensemble-averaged assays[10]. Many biochemical methods have been presented for single-virus assay,such as single-virus multiplexing[11], single-virus genomics[12], and single-virus tracking[13]. However,these methods require biochemical pretreatments(such as extracting viral genomes[11]and labeling viruses with fluorescein[12-13]) on the viruses, which inevitably destroy the integrity of viruses. Compared with these methods, AFM has the unique merit that it is able to visualize the fine structures and measure the nanomechanical properties of single viruses in their native states without any pretreatments. AFM has been widely applied in the studies of individual viruses, such as viral imaging[14], viral nanoindentation[15], viral manipulations[16], and viral capsid self-assembly dynamics[17], contributing much to the emerging field of physical virology[18-19]. By the way, viral physics plays an important role in the life cycle of viruses, and studies have shown that the physical properties of viruses determine how viral genomes traffic in cells, how viruses enter host cells,and how viral genomes are uncoated from the viruses[20]. Hence, investigating the mechanical properties of single viruses is crucial for deciphering the mysteries of viral activities. Despite the wide applications of AFM in virus-related studies, the detailed behaviors of single viruses in liquids are still not fully understood. Particularly, a new AFM imaging mode, which is called peak force tapping(PFT)[21], has emerged in recent years. PFT is a potent mode which allows simultaneously acquiring structural and mechanical images of specimens, and thus PFT is also referred to as multiparametric imaging[22]. PFT-based multiparametric AFM imaging has been utilized to various biomolecular and cellular systems[23-24], but the studies of applying PFT-based multiparametric AFM imaging to viruses are still scarce to the best of our knowledge.
In this work, PFT-based multiparametric AFM imaging was utilized together with AFM indentation assay to investigate the physical behaviors of single virus particles under aqueous conditions. Particularly,the effects of air-drying and alcohol treatment on viral structures and mechanical properties were revealed.The study improves our understanding of viral activities at single-virus level, which will benefit the studies of physical virology.
1 Materials and methods
1.1 Viruses and reagents
The virus particles used here are lentivirus,which were purchased from the OBiO Technology Corporation (Shanghai, China). The virus particle titer was 2.88×108TU/ml. The poly-L-lysine solution was purchased from the Solarbio Life Sciences (Beijing,China). The alcohol (75%) was purchased from the Liaohe Chemical Plant (Shenyang, China). The pure water (Milli-Q, Merck KGaA Company, Darmstadt,Germany) was used in this study.
1.2 Sample preparation
The purchased virus particle solution was diluted 10 times with pure water, and the diluted virus particle solution was used in the subsequent experiments. The purchased poly-L-lysine solution was diluted 10 times with pure water, and the diluted poly-L-lysine solution was used to treat the coverslips to immobilize virus particles for AFM experiments. A fresh coverslip was placed in the diluted poly-L-lysine solution for 24 h, after which the coverslip was airdried. The virus particle solution was then dropped onto the poly-L-lysine-treated coverslip and incubated for 5 min. The coverslip was then washed by pure water to remove the unbound virus particles.Subsequently, the coverslip was placed in a petri dish containing pure water. Next, the petri dish was placed on the sample stage of AFM and individual virus particles were probed by AFM in pure water (Figure 1a). For imaging virus particles in air, the virus particles were attached onto the poly-L-lysine-coated coverslip as described above, and then the coverslip was directly placed on AFM’s sample stage and AFM images were obtained in air. In order to examine the effects of alcohol, the virus particles were attached onto the poly-L-lysine-treated coverslip and incubated for 5 min. Subsequently, the alcohol solution (75%)was added onto the coverslip and incubated for 1 min or 5 min. After that, the alcohol solution on the coverslip was removed by a micropipette. Next, the coverslip was placed in a petri dish containing pure water, which was then placed on the sample stage of AFM and AFM experiments were performed in pure water.

Fig. 1 Experimental platform of characterizing single native virus particles by AFM under aqueous conditions
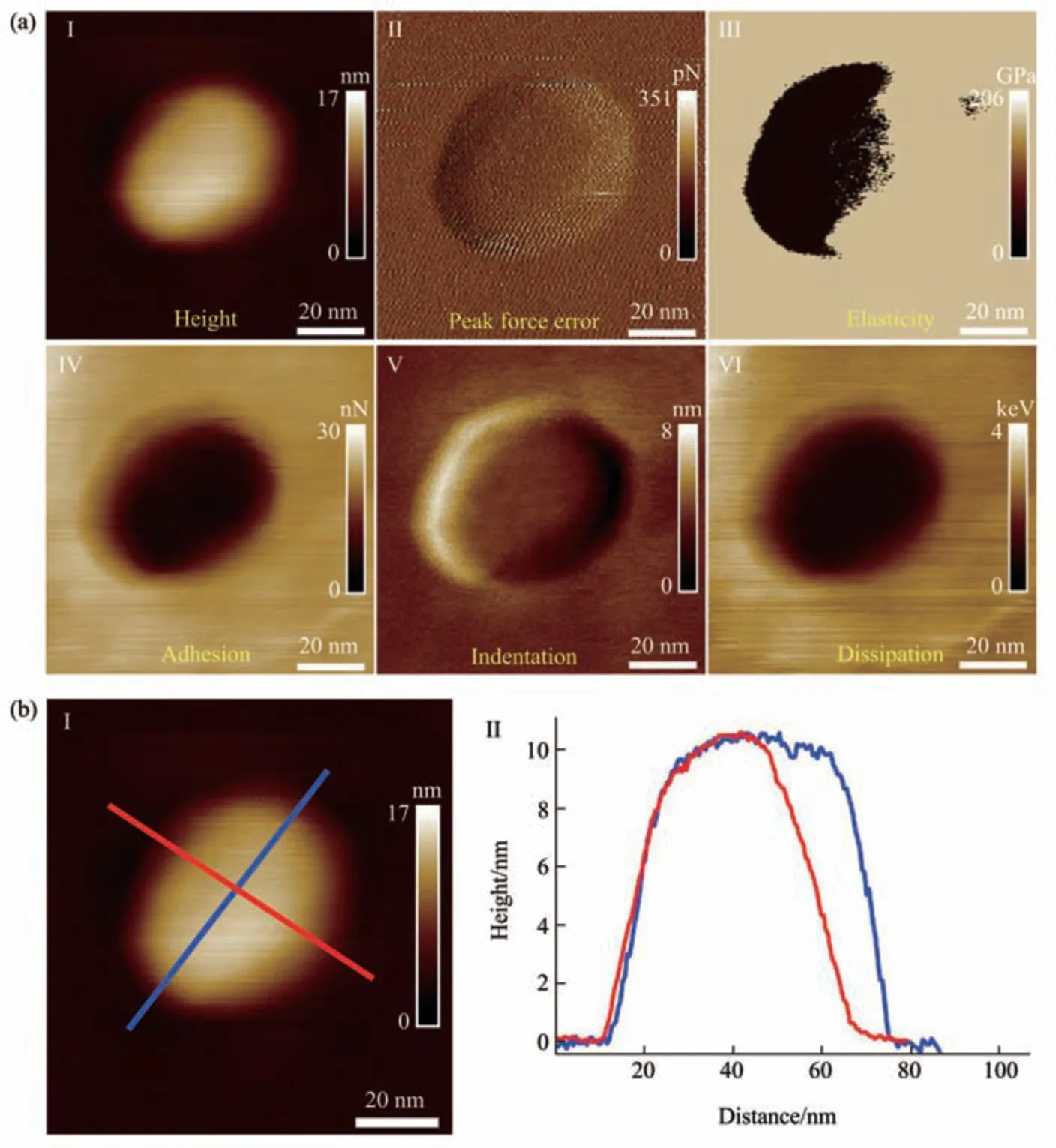
Fig. 2 Multiparametric AFM imaging results of a virus particle obtained in air
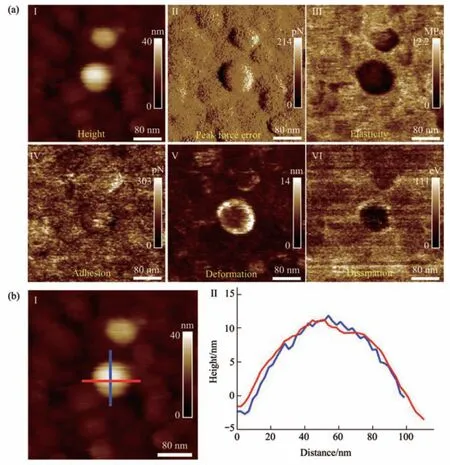
Fig. 3 Multiparametric AFM imaging results of a virus particle obtained in pure water
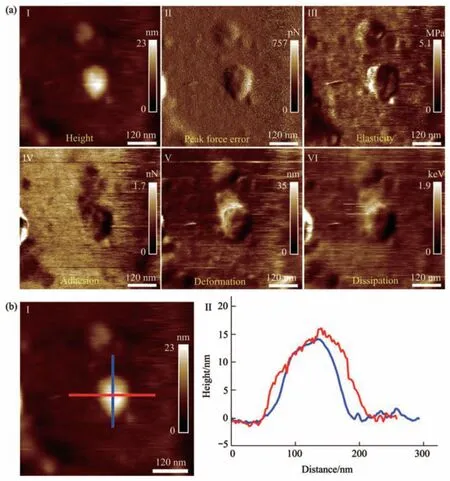
Fig. 4 Multiparametric AFM imaging results of a virus particle treated by alcohol for 1 min
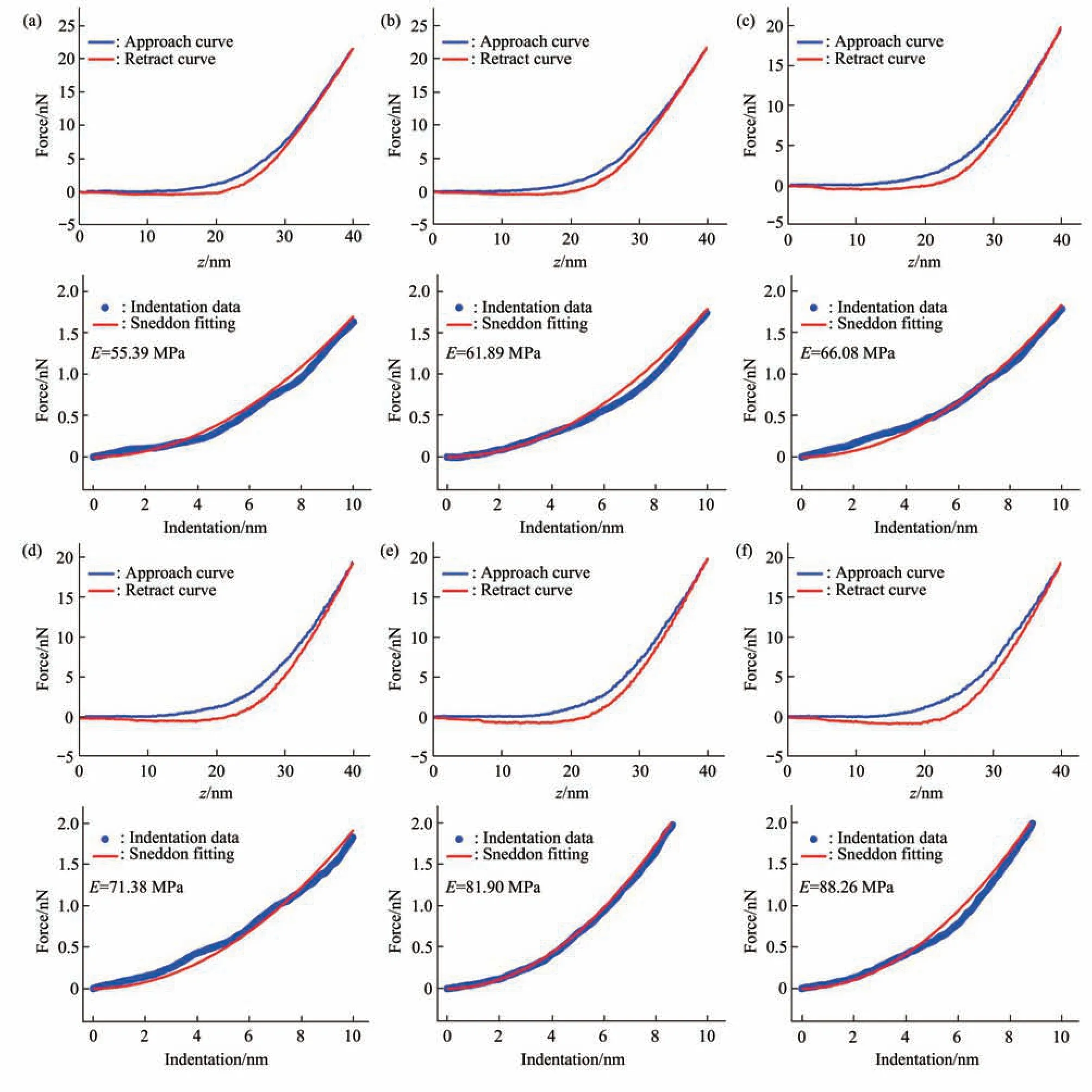
Fig. 5 Typical force curves obtained at different ramp rates on a virus particle without alcohol treatment in pure water
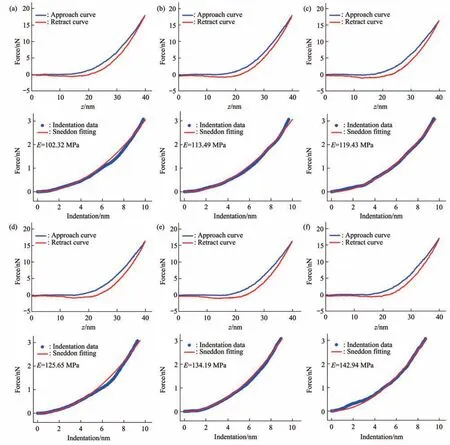
Fig. 6 Typical force curves obtained at different ramp rates on a virus particle treated by alcohol for 1 min
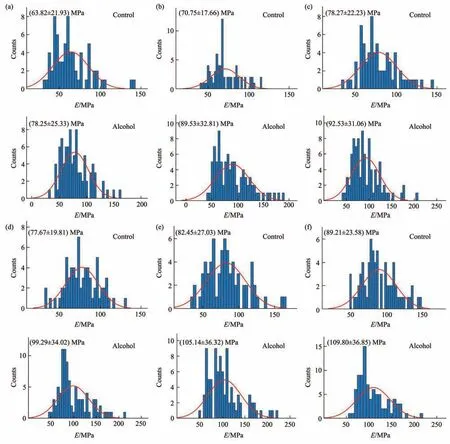
Fig. 7 Statistical histograms of the Young’s modulus changes of virus particles treated by alcohol for 1 min
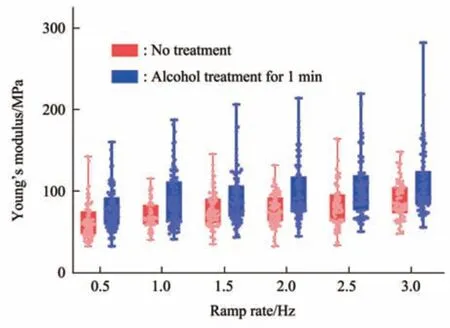
Fig. 8 Box plots of the Young’s modulus changes of virus particles treated by alcohol for 1 min
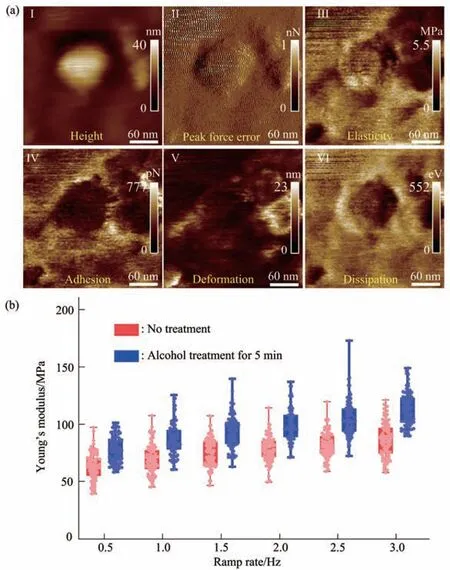
Fig. 9 Multiparametric AFM imaging and Young’s modulus changes of single virus particles treated by alcohol for 5 min
1.3 AFM imaging and indentation assay
AFM imaging and mechanical measurement experiments of virus particles were performed with the commercial Dimension Icon AFM (Bruker, Santa Barbara, CA, USA), as shown in Figure 1b. The AFM has an optical microscope (Figure 1c), which allows moving AFM probe to target areas on the substrate under the guidance of optical microscopy (Figure 1d).The AFM probes used in this study were also purchased from Bruker Company (Santa Barbara, CA,USA). The type of AFM probe used for imaging under aqueous conditions is ScanAsyst-Fluid, whose nominal spring constant is 0.7 N/m and the nominal tip radius is 20 nm (Figure 1d). The type of AFM probe used for imaging in air is ScanAsyst-Air, whose nominal spring constant is 0.4 N/m and the nominal tip radius is 2 nm. Both of the two types of probes are made of silicon nitride with a cantilever shape of triangle. After the blind engagement of AFM probe onto the substrate, force curves were obtained on the bare area of the substrate to calibrate the deflection sensitivity of the probe, which was then used to calibrate the exact spring constant of the probe by using AFM’s thermal noise module. For imaging virus particles, the large-size (2×2 μm2) scanning at PFT mode was performed firstly to find virus particles, and then small-size (100×100 nm2or 600×600 nm2) scanning at PFT mode was performed on individual virus particles. The scan rate was 1 Hz, and 256×256 pixel images were recorded. From the AFM images of single virus particles, the AFM probe was controlled to move to the central area of single virus particle to perform indentation assay and force curves were obtained. For each virus particle, force curves were obtained at six different ramp rates (0.5, 1, 1.5,2, 2.5, 3 Hz) of AFM probe and 10 force curves were obtained at each ramp rate.
1.4 Data analysis
Since the shape of AFM tip used in the study is conical, the Sneddon-modified Hertz model was used to extract the Young’s modulus of virus particles from the obtained force curves[25]:
whereυis the Poisson ratio of virus (hereυ=0.5),Fis the loading force exerted by AFM probe,δis the indentation depth,Eis the Young’s modulus of virus,andθis the half-opening angle of conical tip. The loading forceFcould be obtained from the cantilever deflection by Hooke’s law:
wherekis the spring constant of the probe cantilever,andxis the deflection of probe cantilever. The recorded force curves contain the cantilever deflection(x) and the vertical movements (d) of AFM piezoelectric tube. According to the contact point in the approach part of the force curve, the approach curve was converted into the indentation curve by subtracting the cantilever deflection (x) from the vertical movement of the probe(d)[26]. Fitting the indentation curves with the Sneddon model gives the Young’s modulus of virus particles. The fitting was performed with the program written with Matlab(MathWorks, Natick, Massachusetts, USA).
2 Results and discussion
The PFT-based multiparametric AFM imaging was firstly utilized to examine the effects of air-drying on the structure and mechanics of single virus particles. In PFT mode, AFM tip is controlled to approach to and retract from the sample in a pixel-bypixel manner to record force curves with a positional accuracy of ~0.2 nm and forces at piconewton sensitivity, and analyzing the force curves gives multiple mechanical properties (e.g., deformation,adhesion, elasticity, energy dissipation) of the sample together with the topographical images[27]. Figure 2 shows the multiparametric AFM images of a virus particle recorded in air. We can clearly see the ellipsoid structure of the virus particle from the AFM images (Figure 2). Besides the topographical images(I and II in Figure 2a), the virus particle is also discriminable from the adhesion image (IV in Figure 2a), indentation image (V in Figure 2a), and energy dissipation image (VI in Figure 2a). However, the quality of elasticity image is quite poor and we cannot obtain the elasticity image of the whole virus particle(III in Figure 2a). Section curves (II in Figure 2b)taken along the height image of the virus particle (I in Figure 2b) show that the height of the virus particle is about 10 nm, the size of the long axis of the virus particle is about 70 nm, and the size of the short axis of the virus particle is about 65 nm. Figure 3 shows the multiparametric AFM images of a virus particle recorded in pure water. The substrate was coated with poly-L-lysine biomolecules which are positively charged, allowing attaching the negatively charged virus particles onto the substrate for AFM imaging.From AFM images (Figure 3a), we can clearly see the spheroid structure of the virus particle. Section curves(Figure 3b) show that the height of the virus particle is about 12 nm, and the diameter of the virus particle is about 100 nm. We can see that although the height of the virus particle in aqueous condition is similar to that in air, but the diameter of the virus particle in aqueous condition is significantly larger than that in air, suggesting that the virus particle could shrink in air due to air-drying. Besides, the elasticity image (III in Figure 3a) of the virus particle in aqueous conditions can be well obtained by multiparametric AFM imaging, whereas it is difficult to obtain the elasticity image of the virus particle by multiparametric AFM in air (III in Figure 2a),suggesting that performing AFM experiments in liquids facilitates better characterizing the mechanical properties of virus particles. An interesting phenomenon is that the virus particle is significantly discriminable in elasticity image (III in Figure 3a),deformation image (V in Figure 3a), and dissipation image (VI in Figure 3a), but the contrast between virus particle and the substrate is weak in the adhesion image (IV in Figure 3a), indicating that the adhesive properties of the virus particle are similar to that of the substrate in pure water. In contrast, the adhesive properties of virus particles are quite different from that of the substrate in air (IV in Figure 2a),suggesting the effects of environments (in air or in liquid) on the adhesion of virus particles.Multiparametric AFM imaging has been widely applied to investigate the behaviors of various biological systems, such as native membranes[28],native exosomes[29], native hydrogels[30], living microbial cells[31], and living animal cells[32].However, studies of utilizing multiparametric AFM imaging to probe the activities of native viruses are still scarce. Here, our results show the exciting capabilities of multiparametric AFM imaging in detecting the structures and mechanics of single native virus particles under aqueous conditions, which will benefit investigating the relationship between viral structures and viral mechanical properties at single-virus level.
Multiparametric AFM imaging was then applied to examine the structural and mechanical changes of single native virus particles in response to chemical treatments. Here, alcohol solution was used as an example of chemical stimulus. Alcohol is an effective sanitizer used widely, and viruses are often inactivated in several minutes after being treated by alcohol[33].Traditional studies of viral inactivation are performed on ensemble viruses to reveal the biochemical changes of viruses induced by virucidal agents[34],which are unable to unveil the detailed changes of single viruses during the process of inactivation. With the use of multiparametric AFM imaging, the structural and mechanical alterations of single viruses during the process of inactivation can be clearly visualized. Figure 4 shows the multiparametric AFM images of a virus particle recorded in pure water after being treated by alcohol solution (75%) for 1 min. We can see that the virus particle can be identified from both topographical images (I and II in Figure 4a) and mechanical images (III-VI in Figure 4a). However,compared with the virus particles without alcohol treatment (Figure 3), mechanical changes of the virus particles after alcohol treatment could be remarkably observed. For example, the deformation property of the virus particle significantly weakened after alcohol treatment (V in Figure 4a), whereas the virus particle without alcohol treatment had strong deformation capability (V in Figure 3a). Besides, the elasticity distributions of the virus particle after alcohol treatment are quite heterogeneous (III in Figure 4a),while the elasticity distributions of the virus particle without alcohol treatment are homogeneous (III in Figure 3a). In addition, we can see that the shape of the virus particle without alcohol treatment is more like a ball (Figure 3a), while the shape of the virus particle after alcohol treatment becomes irregular(Figure 4a). Figure 4b shows the geometric features of the virus particle, which are close to that of the virus particle without alcohol treatment (Figure 3b).Overall, the results show that the effects of alcohol treatment on the structures and mechanical properties of single virus particles can be clearly visualized by multiparametric AFM imaging.
Since multiparametric AFM imaging only qualitatively shows the effects of alcohol treatment on virus particles, we then used conventional AFM indentation assay to quantitatively characterize the effects of alcohol treatment on the mechanical properties of single virus particles. After locating the single virus particles from the topographical images obtained by AFM imaging, AFM probe was moved to the center of virus particle to obtain force curves.Figure 5 shows the typical force curves obtained on a virus particle without alcohol treatment at different ramp rates (the ramp rate indicates the loading velocity of the AFM probe during indentation assay)together with the theoretical fitting results of calculating the Young’s modulus of the virus particle.After converting the approach curve into the indentation curve, the Young’s modulus of the virus particle was obtained by fitting the indentation curve with Sneddon-modified Hertz model. We can see that the experimental indentation curves are quite consistent with the Sneddon fitting curves, suggesting the effectiveness of characterizing the indentation process on the virus particle by Sneddon-modified Hertz model. Figure 6 shows the typical force curves obtained on a virus particle treated by alcohol for 1 min and the theoretical fitting results, and we can also see that the indentation process on the virus particle is well characterized by the Sneddon model.Notably, for utilizing AFM indentation assay and Hertz-Sneddon model to characterize the mechanical properties of living cells, the indentation depth should be less than 10% of the cell thickness to avoid the influence from the stiff substrate beneath the cell[35].Here, we can see that the Sneddon model is still applicable even though the indentation depth is larger than 10% of the virus thickness (Figure 5, 6), and this may be due to the structural characteristics of the viruses. It is well known that the structures of viruses are much simpler than that of cells[36], which may therefore facilitate theoretically modeling the mechanical behaviors of the viruses. The statistical histograms of the Young’s modulus values of virus particles before and after alcohol treatment were then obtained, as shown in Figure 7. The box plots of the results in Figure 7 are shown in Figure 8. From the statistical results (Figure 7, 8), we can see that the Young’s modulus of the virus particle increases with the ramp rates of AFM probe. For the indentation assay, the mechanical properties of biological systems depend on the loading rate (the force increasing over time) at which they are measured, and thus it is meaningless to compare the mechanical properties of biological systems without specifying the loading rate[37]. Studies have shown the Young’s modulus of cells measured by AFM increases with the loading rates[38-39]. Here, we can also see that the Young’s modulus of virus particles measured by AFM indentation assay increases with the loading rates.Measuring the mechanical properties of viruses at different loading rates therefore benefits understanding the dynamic mechanical behaviors of viruses. Besides, the indentation assay results (Figure 7, 8) show that virus particle significantly stiffens after the alcohol treatment. With the established method, we then visualized and measured the mechanical dynamics of virus particles treated by alcohol for 5 min, and the results are shown in Figure 9. From the multiparametric AFM images, we can clearly see that the stiffness of the virus particle significantly increases (III in Figure 9a) and accordingly the deformability capability of the virus particle significantly decreases (V in Figure 9a). An interesting point is that, unlike the virus particles without alcohol treatment, the virus particles become discriminable in the adhesion images after the alcohol treatment (IV in Figure 9a and IV in Figure 4a),suggesting the changes of the adhesion of the virus particles caused by alcohol. AFM indentation assay also quantitatively shows the increase of Young’s modulus of virus particles after the alcohol treatment(Figure 9b), which are consistent with the results for the virus particles treated by alcohol for 1 min (Figure 8). Overall, these results provide direct evidence for the effects of alcohol treatment on viral mechanics.
The study presented here will have general implications for the studies of virology from the perspective of single-virus mechanics. Mechanics is essential for the life cycle of virus, since viruses should be sufficiently mechanically stable to protect their genome in the extracellular environment, but they also should be sufficiently unstable that they can release their genomic materials into host cells[18].Therefore, revealing the detailed mechanical cues involved in the activities of viruses is crucial for exploiting viruses or virus-like particles to fight against human diseases[40]. However, so far, the role of viral mechanics in viral life cycle is still poorly understood. Here, the experimental results remarkably show the capabilities of multiparametric AFM imaging in visualizing single native virus particles and their structural and mechanical dynamics in response to chemical stimuli under aqueous condition(Figure 2-4, 9), providing a novel way to simultaneously characterize the structures and mechanical properties of single viruses. The AFM indentation experiments (Figure 5-9) reveal the significant changes of viral mechanics after the treatment of alcohol, improving our understanding of the viral mechanics. Notably, the virus used here is lentivirus which belongs to the enveloped viruses.Researchers have used AFM to investigate the nonenveloped viruses such as the adenovirus[41-42], and thus utilizing multiparametric AFM imaging to investigate the mechanical behaviors of nonenveloped viruses will promote exploring more viral activities. Overall, the results obtained by AFM significantly complement traditional ensembleaveraged assays, allowing understanding the detailed behaviors of single viruses in their native states.Particularly, the established methods (e.g., virus immobilization in aqueous condition, multiparametric AFM imaging, AFM indentation assay) presented here can be directly applied to other virus systems,which will bring novel insights into the pathogenesis of viruses and will potentially benefit developing novel strategies for antiviral therapy.
3 Conclusion
The study provides a novel way to characterize the structural and mechanical behaviors and their dynamics of single native viruses under aqueous conditions based on AFM, which will benefit the studies in the field of physical virology and will be meaningful for uncovering the underlying mechanisms guiding the mysteries of viral activities.
- 生物化學與生物物理進展的其它文章
- 探究式課程設計在醫(yī)學生臨床決策能力培養(yǎng)中的探索*
- 基于徑向基函數(shù)神經(jīng)網(wǎng)絡的肺部加權(quán)頻差電阻抗成像方法*
- 高內(nèi)涵分析系統(tǒng)聯(lián)合激光掃描共聚焦顯微鏡觀察細胞間隧道納米管*
- 胃容積及胃食道液體狀態(tài)變化的電學特性研究*
- Transcriptomic Analysis of Deinococcus radiodurans During The Early Recovery Stage From Ultraviolet Irradiation*
- 玉竹提取物對大鼠多囊卵巢綜合癥的療效研究*