Advancing approach and toolbox in optimization of chloroplast genetic transformation technology
LIU Yu-xin,LI Fan,GAO Liang,TU Zhang-li,ZHOU Fei#,LIN Yong-jun
1 National Key Laboratory of Crop Genetic Improvement/College of Life Science and Technology,Huazhong Agricultural University,Wuhan 430070,P.R.China
2 Hubei Hongshan Laboratory,Wuhan 430070,P.R.China
3 Wuhan Towin Biotechnology Co.,Ltd.,Wuhan 430070,P.R.China
Abstract Chloroplast is a discrete,highly structured,and semi-autonomous cellular organelle. The small genome of chloroplast makes it an up-and-coming platform for synthetic biology. As a special means of synthetic biology,chloroplast genetic engineering shows excellent potential in reconstructing various sophisticated metabolic pathways within the plants for specific purposes,such as improving crop photosynthetic capacity,enhancing plant stress resistance,and synthesizing new drugs and vaccines. However,many plant species exhibit limited efficiency or inability in chloroplast genetic transformation. Hence,new transformation technologies and tools are being constantly developed. In order to further expand and facilitate the application of chloroplast genetic engineering,this review summarizes the new technologies in chloroplast genetic transformation in recent years and discusses the choice of appropriate synthetic biological elements for the construction of efficient chloroplast transformation vectors.
Keywords: chloroplast,genetic engineering,new technology,plasmid optimization,nanotechnology
1.Introduction
It has been estimated that the world’s population will exceed nine billion in 2050,and the global demand for food will increase by 100-110% compared with that in 2005 (Tilmanet al.2011). The traditional farming model is overall cumbersome and inefficient. With the rapid population growth,it is a great challenge to meet the global demand for food in future agricultural production with limited land resources. Compared with other approaches,synthetic biology is considered the most promising approach for future agricultural production through the genetic improvement of crops and the construction of efficient metabolic pathways.Several notable breakthroughs in synthetic biology have been achieved in microbial systems,which have been gradually extended to eukaryotes and even plants.With the construction of metabolic pathways,plant genetic transformation is an efficient means for crop improvement,in which nuclear genome engineering is the most widely used for the transfer of desired genes into the nuclear genomes of cultivated species. However,further development and application of this technology have been largely hindered by the possible transgene diffusion caused by the hybridization of nuclear transgenic plants with wild relatives (Miroshnichenkoet al.2016;Divelyet al.2020). Maternal inheritance of chloroplasts in higher plants may present a potential strategy to address the issue of biosafety. Moreover,compared with nuclear genetic transformation,chloroplast genetic transformation uses chloroplasts as a storage space for specialized proteins so as to circumvent their toxicity through accumulation in the cytoplasm (Daniellet al.2001,2005). The successful chloroplast transformation inChlamydomonas reinhardtii(Boyntonet al.1988)andNicotiana tabacumL.(Svabet al.1990) marked the beginning of plant chloroplast engineering. With the progress in laboratory research on chloroplast genetic engineering around the world,chloroplast transformation has been successfully achieved,and reliable transformation systems have been established for more than 20 species of flowering plants,most of which are dicotyledonous plants (Table 1).
Chloroplast genetic engineering comprises the modification of genomes and the introduction of foreign genes. A mature and stable chloroplast genetic system involves effective transformation methods and the design of transformation vectors with correct selection markers,regulatory elements,and insertion sites. A chloroplast genetic transformation system cannot be completely shared by different species. On the one hand,the transformation efficiency varies greatly with different chloroplast transformation methods (Zienkiewiczet al.2017). On the other hand,the design of a suitable transformation vector with selection markers and expression elements can greatly improve the success rate of chloroplast genetic transformation. Over the past years,numerous novel approaches have been applied to overcome the obstacles in chloroplast genetic engineering.
This review summarizes the newly emerging technologies in chloroplast genetic transformation in recent years (Fig.1;Table 2). It also discusses the key elements for the design of chloroplast transformation vectors to enhance the transformation efficiency.
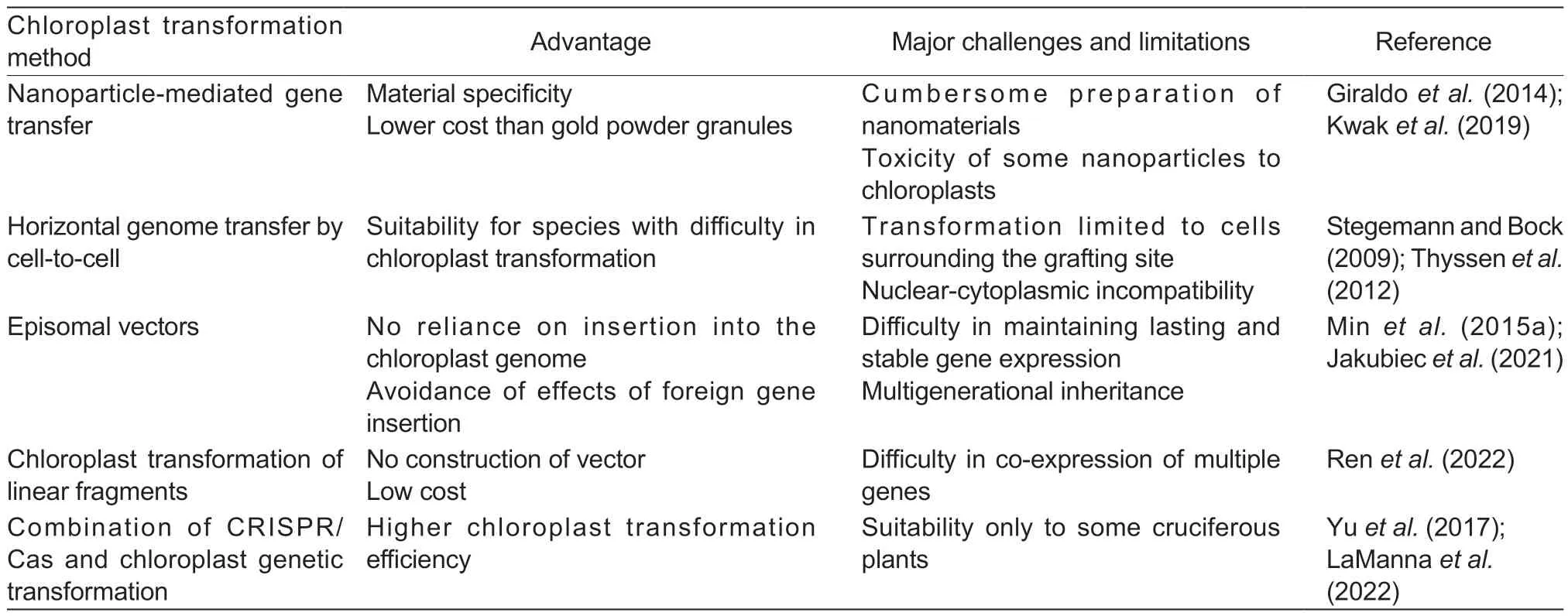
Table 2 Advantages and challenges of new technologies for chloroplast transformation
2.New approaches for DNA delivery
Different fromAgrobacterium-mediated nuclear transformation,chloroplast genetic transformation mostly relies on physical or chemical methods. Biolistic bombardment is the most extensively used method for the transfer of exogenous genes into recipient plants,whose basic principle is to bombard tiny particles(tungsten or gold particles with diameters of 0.6-1.6 μm)attached with foreign genes into the chloroplasts of the recipient plant by high-pressure helium in a vacuum-like environment. This method is not limited to any specific species or plant cells,but the high-pressure penetration of tiny particles will inevitably cause damage to cells(Kumar and Ling 2021). In addition,polyethylene glycol(PEG)-mediated transformation has been applied in chloroplast transformation. By taking plant protoplasts as the recipient cells,exogenous DNA is facilitated to pass through the cell membrane to enter into the chloroplast and is then integrated into the genome in the form of vesicles under the mediation of PEG (Yuet al.2020). However,this method is largely limited by the low regeneration efficiency of the transformed protoplast,which is highly dependent on the species and transformed tissue. In addition,there are some other transformation methods,such as glass bead transformation (Economouet al.2014;Wannathonget al.2016) and microinjection method (Knoblauchet al.1999). However,these methods have not become mainstream methods due to various reasons such as instability of the vector,limitation of transformed species,and low expression efficiency.Therefore,more effective tools are needed to extend the applicability of chloroplast genetic engineering.
2.1.Nanoparticle-mediated transfer
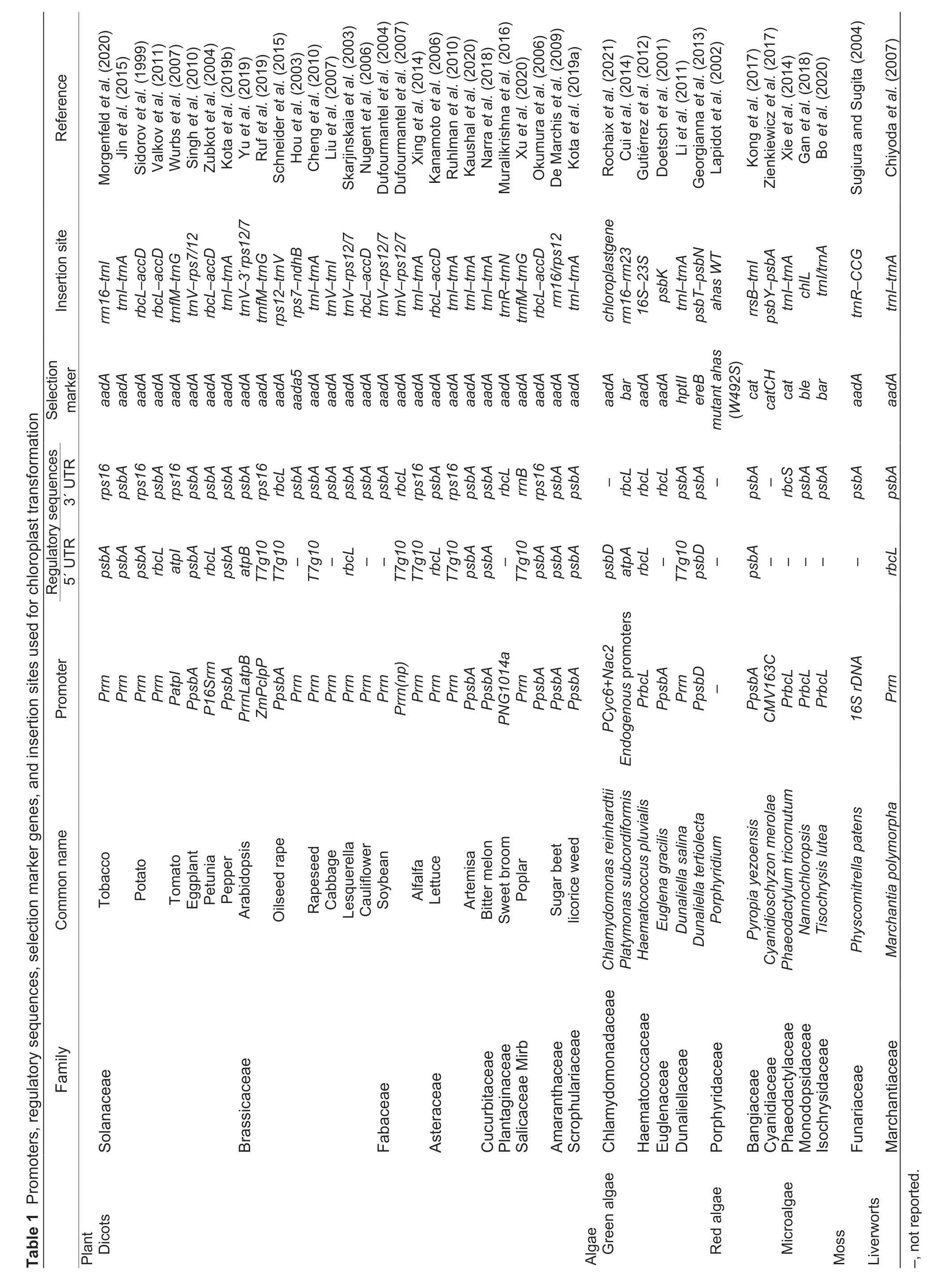
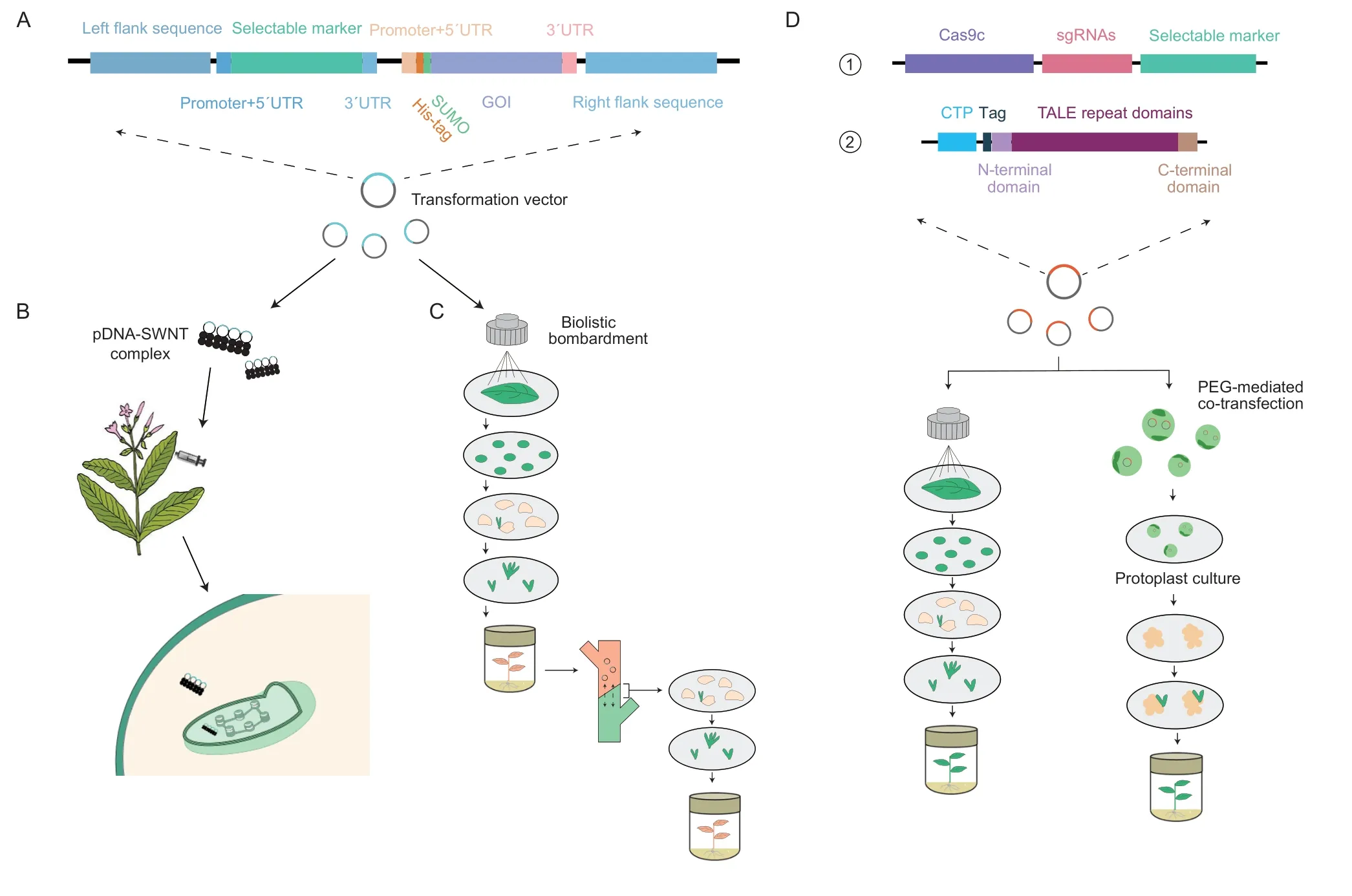
Fig. 1 Schematic diagram of transformation plasmid and novel transformation method. A,schematic diagram of transformation plasmid. B,process of genetic transformation mediated by nanoparticles. C,the transformation process of cell-to-cell horizontal genome transfer. D,①,schematic diagram of CRISPR/Cas transformation plasmid;②,the structure of TAL effectors can be transformed by biolistic bombardment and PEG-mediated transformation. GOI,gene of interesting;SUMO,small ubiquitin-like modifier;CTP,chloroplast transit peptide.
In the establishment of precise chloroplast transformation systems applicable to multiple species,nanomaterials have shown revolutionary potential in chloroplast genetic engineering owing to their unique physicochemical properties. Nanotechnology,as an emerging plant molecular tool,has numerous applications,such as the improvement of stress tolerance (Djanaguiramanet al.2018;Wuet al.2018),material transport efficiency(Wonget al.2016),metabolite yield (Zhanget al.2022),photosynthesis (Namet al.2010;Giraldoet al.2014;Chowdhuryet al.2018),and creation of new plant signaling molecules and pollutant detectors(Giraldoet al.2014;Komanet al.2017). In the field of DNA transformation,silicon carbide (SiC) fibers were successfully used in the genetic transformation of plants for the first time in 1990 (Kaeppleret al.1990). The wear of cell barrier caused by SiC fibers indicates the potential of nanoparticles to deliver exogenous DNA into organelles. It has been further demonstrated that encapsulated foreign DNA can be delivered by nanoparticles from various raw materials (e.g.,silicon,gold,carbon,and cerium) through plant cell walls and membranes and then expressed in cells,and the delivery efficiency can be improved by manipulating the aggregate shape,aspect ratio,and surface charge(Demireret al.2019;Huet al.2020). However,the intracellular localization of nanoparticles is mostly random in the cytoplasm,and only some nanoparticles can be internalized into chloroplasts (Demireret al.2019). Therefore,it remains highly challenging to deliver nanoparticle-mediated genes to specific organelles in mature plants. Previous studies have revealed that ordinary single-walled carbon nanotubes (SWNTs) can penetrate plant cell walls,membranes,and even the bilayer lipid membrane of chloroplasts,but they cannot be precisely localized in the chloroplast (Giraldoet al.2014). Chitosan-wrapped SWNTs (CS-SWNTs) can be precisely localized in chloroplasts at sufficiently high surface charges (Giraldoet al.2014;Kwaket al.2019).The primary amines of CS-SWNTs have a pKaof 6.5,below which the amine groups can be protonated and strongly bind to plasmid DNA (pDNA). In the designed CS-SWNTs-DNA complexes,DNA was tightly bound to CS-SWNTs in the weakly acid environment,such as the plant cytoplasm (pH~5.5),and the DNA would be released in the weakly alkaline environment,such as the chloroplast matrix (pH~8). With the pH gradients in plant cells,CS-SWNTs can specifically transport pDNA into the chloroplasts of different plant species(such as mustard,watercress,tobacco,spinach,andArabidopsis) for transient expression. Simultaneously,as a biodegradable polysaccharide,chitosan is of no toxicity to plants. In addition,negatively charged poly (acrylic acid) nanoceria (PNC) can also be transported into chloroplasts in mesophyll cells through a non-endocytic pathway,and the co-localization rate of chloroplasts is as high as 46% (Wuet al.2017). A recently reported novel engineered nanotechnology utilizes a peptide recognition motif to transport a hydrophilic quantum dot (QD) into the chloroplast. The QD is encased in a molecular basket of β-cyclodextrin,whose size allows the transport of QD through leaf cell wall pores. By using the highly conserved Rubisco small subunit 1A (RbcS)in multiple species as guide peptides,the complexes can bypass the biological barriers of plants and deliver chemicals to chloroplasts (Santanaet al.2020). Although nanotechnology has developed rapidly,foreign expression in transgenic plants obtained by those methods has only been verified by the cell biology approach. There is still a long way to go to integrate nanotechnologies into plant genetic approaches.
2.2.Cell-to-cell horizontal genome transfer
Transmission of genetic materials between species mostly occurs through sexual reproduction,that is,the exchange of genetic materials through hybridization.However,recent genetic experiments have revealed that the nuclear,chloroplast,and mitochondrial genomes can be horizontally transferred asexually between cells of different species (Stegemann and Bock 2009;Rebbecket al.2011;Thyssenet al.2012;Fuenteset al.2014;Tanet al.2015;Gurdonet al.2016;Bock 2017). Unlike that of nuclear and mitochondrial genes,the horizontal transfer of the chloroplast genome can be achieved by grafting (Bock 2017),and the mechanism is similar to the formation of plasmodesmata between the parasitic plant and the host(Melnyk and Meyerowitz 2015). When the species is grafted,the cell wall structure of cells around the grafting site will change significantly to form larger stomata,and the chloroplast is also transformed into the shape of a fluid amoeba to ensure that it can pass through the stomata between cell walls and enter another cell (Hertleet al.2021). However,the transformation only occurs in those cells surrounding the grafting site (Stegemann and Bock 2009).
Horizontal transfer of genome between cells provides new implications for the transformation of species with difficulty in chloroplast transformation. For example,Nicotiana glauca,a nicotine-free,fast-growing,and highbiomass-yielding tobacco variety,shows higher efficiency thanN.tabacumin exogenous gene expression and is therefore considered a potential biosynthetic factory(Ahrazemet al.2022). In recent years,transgenic materials accumulating crocin and ketocarotenoids inN.glaucahave been obtained by nuclear transformation engineering (Mortimeret al.2017;Ahrazemet al.2022). However,no chloroplast transformation scheme suitable forN.glaucahas been available so far. In order to produce high-value ketocarotenoid astaxanthin in the chloroplast genome ofN.glauca,transplastomicN.tabacumwas grafted withN.glauca,and the transgenic chloroplast genome could move across the graft junction fromN.tabacumplants intoN.glauca. Then,seven independent gene transfer lines were obtained through a double selection of spectinomycin and kanamycin. Six of the seven lines were homoplasmic for the transferred chloroplast genome fromN.tabacum,and only one line was heteroplasmic with a mix of wild-typeN.glaucaand transgenicN.tabacumchloroplast genomes (Luet al.2017).
In addition to the above-mentioned formation of plasmodesmata between cells by grafting,a new technique called “Cell Grafting” was developed,which utilizes a mixed population of callus cells from two parental plants grownin vitroto achieve horizontal transfer of chloroplast or nuclear genomes (Sidorovet al.2018). This technique effectively avoids the tedious grafting work and is suitable for the chloroplast transformation of species that cannot be conventionally grafted and monocotyledonous plants with low chloroplast transformation efficiency.
However,it is worth noting that this horizontal transfer of chloroplast genomes may be only applicable to relatively close species. This is because with increasing phylogenetic distance in chloroplast genome between the donor and recipient species,horizontal transfer of chloroplast genomes can lead to nuclear-cytoplasmic incompatibility (Schmitz-Linneweberet al.2005;Stegemannet al.2012).
3.Design of vectors for efficient chloroplast transformation
A hallmark of chloroplast biotechnology is the use of homologous recombination (HR) to insert exogenous genes with selection markers into native chloroplasts,followed by multiple rounds of stress screening to generate stable transplastomic systems. Therefore,a typical chloroplast transformation vector consists of flanking sequences for homologous recombination,inserted foreign genes and specific expression elements (Fig.1-A).
3.1.Choice of flanking sequences: HR-based chloroplast engineering
Integration of the foreign DNA into the chloroplast genomeviaHR is a crucial step after DNA delivery into the chloroplast. To ensure the optimal HR efficiency between the transformation vector and plant plastome,the chloroplast transformation vector needs to carry two flanking regions,which enable insertion of the foreign DNA at any desired site in the chloroplast genome or specifically modify any chloroplast encoded gene of interest. The flanking sequence is chosen based on the insertion site of the foreign gene or the specific gene to be knocked out or modified,and its length is generally 1-1.5 kb.Over the past years,many insertion sites have been identified in the chloroplast genome,and the commonly used insertion sites for foreign gene expression includetrnI-trnA,rbcl/accD,trnV-rps7/12,andtrnfM/trnG. Since there have been few studies comparing the insertion of the same gene at different positions,it is impossible to exactly determine the optimal insertion site (Bock 2014).
3.2.Expression of foreign genes from episomal vectors: An alternative to traditional HR-based chloroplast engineering
It is generally believed that there is no alternative to HR for chloroplast genetic engineering. However,the replication properties of geminiviruses (Jeskeet al.2001) and the small circular chloroplast genome of dinoflagellates (Howeet al.2008) make it possible to develop methods that do not rely on the insertion of DNA into chloroplast genome and only stably amplify foreign DNA in organelles.
Geminiviruses are plant DNA viruses relying on rolling circle replication and recombination-dependent mechanisms to replicate in cells,and the double-stranded intermediates produced by replication can serve as viral protein transcription templates (Gutierrez 1999;Jeskeet al.2001). Therefore,utilization of the replication process of geminiviruses may facilitate the stable amplification of foreign DNA in organelles independent of chloroplast HR. The first attempt to develop a viral episomal vector for chloroplast transformation was carried out based on Rep (replication initiation function)and VOR (viral origin of replication) elements from beet curly top geminivirus (BCTV) (Jakubiecet al.2021).The results showed that the exogenous DNA flanked by VORs in the presence of a chloroplast-localized Rep could be efficiently amplified as a physically independent minichromosome in plant chloroplasts without insertion into the chloroplast genome. This can effectively avoid the position effect from adjacent expression elements(Quesada-Vargaset al.2005;Sharwoodet al.2011),and contribute to the formation of a tightly regulated inducible expression system in chloroplasts.
Different from the single large ring-packed chloroplast genome (107-218 kb) of common angiosperms(Chumleyet al.2006;Linet al.2010),the chloroplast genome of dinoflagellates can be divided into multiple independent small rings (2-3 kb),each of which consists of a coding region and a non-coding region.In most cases,the coding region consists of a single chloroplast gene and a promoter (Zhanget al.1999,2001). This unique form of chloroplast genome can allow the introduction of single foreign genes into plant organelles for independent replication,which is similar to plasmid replication in bacteria. The extrachromosomal DNA of the dinoflagellateH.triquetracontains a putative replication origin that can sustain and replicate in higher plant chloroplasts in the form of an episomal vector carrying transgenes. After the vector is bombarded into tobacco,it can be expressed independently for at least six months without insertion into the chloroplast genome. Unfortunately,interruption of the selection pressure will lead to the rapid loss of the episomal vector (Minet al.2015a). To avoid the effect of selection pressure and achieve long-term expression of the episomal vector,a novel mini-synplastome vector containing episomal plasmid replication components(oriA2andA1and NICE1 (nicotiana chloroplast extrachromosomal element)) was transformed into chloroplasts of potato. It was found that the ratio of the copy number of episomal DNA to that of plastome in genomic DNA could reach 1-2,and the expression efficiency was comparable to that of HR-based chloroplast engineering. In the absence of selection pressure,mini-synplastomes can also be maintained across multiple vegetative generations of plants (Staub and Maliga 1994;Occhialiniet al.2022).
3.3.Selection of regulatory elements
Constitutive promotersPromoter is an important sequence to improve the abundance of transgene mRNA and protein expression level. The appropriate choice of promoters can ensure the function of chloroplast transgene. Higher plant chloroplasts have two different RNA polymerase activities: chloroplast-encoded RNA polymerase (PEP) and nuclear gene-encoded RNA polymerase (NEP). They recognize different types of promoters. The PEP promoter is similar to the bacterial σ70-type promoter and contains -35 (TTGACA) and-10 (TATAAT) regions,while the NEP promoter has a core sequence motif (YRTA) similar to the consensus sequences of promoters in plant mitochondrial genomes(Bock 2021). Generally,the PEP promoter appears to be much stronger than the NEP promoter;therefore,the PEP promoter is primarily used to drive the expression of chloroplast transgenes. At present,the promoter with the highest efficiency in driving gene expression is therrnpromoter,driving the ribosomal RNA operon.Therrnpromoter can result in 3-and 12-fold GFP RNA transcription relative to thepsbApromoter in tobacco chloroplasts and the bacterialtrcpromoter,respectively,and lead to about 90-fold higher GFP protein expression levels than the two promoters (Newellet al.2003). In addition,clpP(Valkovet al.2011),rbcL(Linet al.2003),and the synthetic promoterPNG1014a(Muralikrishnaet al.2016) can also be applied to chloroplast transformation.
Moreover,since the transformation vector takes advantage of HR to insert foreign genes,it should be noted whether the promoter sequence in the transformation vector has homology with the endogenous promoter sequence in the transformed plant. If promoter and terminator sequences homologous to host chloroplast DNA are used for transgene expression,unexpected recombination events may occur,and even extrachromosomal DNA may appear through accidental intramolecular recombination between the homologous promoter and terminator sequences in the vector and chloroplast DNA (Grayet al.2009),which will be retained in progeny (Minet al.2015b). Therefore,currently,researchers are attempting to use promoters from other organisms,such asChlamydomonas reinhardtii,for multiple-gene expression (Fuenteset al.2016;Bock 2022).
Tissue-or development-related promotersMining of new regulatory elements will enhance the chloroplast transformation efficiency of non-green tissues and facilitate the establishment of new transformation systems. An analysis of chloroplast gene expression at different developmental stages of tomato fruit and potato tuber has revealed that except for that of theaccDgene (Kahlau and Bock 2008;Valkovet al.2009),the expression of nearly all non-photosynthesis genes was markedly down-regulated relative to that in green tissues. A recent study investigated the expression levels of chloroplast genes in different tissues of kiwifruit at different developmental stages. The results demonstrated that a few photosynthetic genes (psaI,ycf3,ndhH,andpetG) were up-regulated at the RNA level in fruits compared with those in leaves at early developmental stages,and they maintained high transcription levels in the later stages of fruit development. In contrast,psbAtranscription remained nearly unchanged during chromoplast formation,andrrn16mRNA was nearly equal in leaves and green fruits (Chenet al.2022). These findings facilitate the design of chloroplast expression elements that are expected to overcome the inefficient expression of transgenes in non-green chloroplasts.
Inducible promoters Although constitutive promoters can confer a high-efficiency expression of exogenous genes,the adverse effects of constitutive expression on plants should not be ignored. A seemingly simple biosynthetic pathway may lead to unexpected complex changes,such as developmental abnormality (L?sslet al.2003;Luet al.2017),and even affect plant lifespan(Mageeet al.2004). Due to the lack of endogenous systems for inducible gene expression in chloroplasts,it is necessary to create a regulatory system for chloroplast gene expression. Early inducible systems involve the use of a nucleus-encoded chloroplast-targeted T7 bacteriophage RNA polymerase (T7RNAP) controlled by inducible nuclear promoters and the T7 promoter to construct a transactivation system to drive the expression of the transgene (L?sslet al.2005). In addition,several systems have been developed,including the systems based on thelacregulatory system (Mühlbauer and Koop 2005;Katoet al.2007),and modified sigma factors (Buhotet al.2006). Recently,Rojaset al.(2019) designed the ethanol-inducible orthogonal PPR protein as an activator whose corresponding binding site was placed in the 5′UTR of the transgene. The engineered proteins could stimulate the expression of chloroplast transgenes by up to~40 folds. Although these systems are effective,they also have some obvious disadvantages. For example,the T7RNAP system and PPR regulation require nuclear transformation to assist the protein in entering the chloroplasts,while the LAC operating system requires modification of the promoter sequence,which tends to have unpredictable impacts.
Riboswitches are natural RNA sensors usually located in the non-coding regions of mRNA. When sensing cellular metabolites,riboswitches will undergo some changes in secondary structure and,therefore,can regulate gene expression (Mandalet al.2004;Aghdamet al.2016). Riboswitches are rich in prokaryotes (Winkler and Breaker 2005). Therefore,the semi-prokaryotic nature of chloroplasts makes riboswitches a promising tool to be used as novel gene switches to regulate gene expression in chloroplast genomes. In order to design feasible chloroplast riboswitches,Verhouniget al.(2010) constructed six riboswitch systems. Functional verification inE.coliand tobacco chloroplasts has revealed that although the chloroplasts retain some prokaryotic properties,the regulatory mechanism of gene expression is not exactly the same as that in bacteria.Among them,a synthetic theophylline switch (s.theo-RS)exhibited excellent gene regulation ability inE.coliand tobacco chloroplasts (Verhouniget al.2010).
Although riboswitches can induce gene expression,the transcription efficiency of inducible promoters is usually lower than that of constitutive promoters. In order to improve the transcription efficiency of theophylline riboswitches,an RNA amplification step mediated by bacteriophage T7 RNA polymerase was added to the theophylline riboswitch to form a new RNA amplification enhanced riboswitch (RAmpER). The induced low-level expression of T7 RNA polymerase can trigger strong transcription of the foreign gene (Emadpouret al.2015).Recently,RAmpER was also successfully used to regulate the synthesis of astaxanthin (Agrawalet al.2022),as well as antimicrobial peptides (AMPs) (Hoelscheret al.2022) in tobacco. Transplastomic plants controlled by the RAmpER system showed markedly improved growth phenotypes in the non-induced state compared with under strong constitutive expression.
Selection of 5′ UTRUntranslated region (UTR) plays an important role in the post-transcriptional regulation of gene expression,affecting mRNA transport and subcellular localization (Mignoneet al.2002;Mayr 2019).Clarifying the function of UTR is of great significance for improving the efficiency of chloroplast genetic transformation and protein expression. According to the theory of endosymbiosis,the transcription and translation of chloroplast genes are similar to those of bacterial genes. Evaluation of the chloroplastin vitrotranslation system confirmed that the 5′ UTR of chloroplast mRNA plays an important role in regulating gene expression,just like in bacteria,carrying translation initiation signals and determining mRNA translation rate (Hirose and Sugiura 1996,1997;Kuroda and Maliga 2001;Yukawaet al.2007;Scharffet al.2017). During the initiation of chloroplast mRNA translation,ribosome recruitment depends on the complementarity of the Shine Dalgarno(SD) sequence (GGAGG) of the mRNA to the reverse SD sequence at the 3′ end of the 16S RNA. However,not all chloroplast mRNAs have SD sequences. For example,the 5′ UTR region ofpsbNlacks SD sequence but has two processing sites (-39 and -24). Different processing will affect the translation efficiency ofpsbN.Processing at -39 could enhance the translation rate by 5 folds. The reason for the higher efficiency is that the 18-nt region between the -24 and -7 sequences is the protein factor binding site required for the translation(Kuroda and Sugiura 2014). Therefore,the selection and modification of 5′ UTR are important ways to improve the translation efficiency of transgenic proteins. In the comparative studies of 5′ UTR efficiency (Herzet al.2005;Yanget al.2013),the 5′ UTR of phage T7 gene 10 (T7g10) was identified as the strongest translation initiation signal. The leader sequence ofT7g10contains a perfect SD sequence and can drive an extremely high mRNA translation rate inE.coli. When usingrrnas the promoter,combined with the 5′ UTR ofT7g10for chloroplast transformation,a higher accumulation level of exogenous protein could be obtained (Kuroda and Maliga 2001;Xuet al.2020).
In addition,researchers have developed an inducible repressible and reversible expression system inChlamydomonas,which is based on the finding that Nac2 protein is an essential protein factor for the stable translation of the chloroplast genepsbDmRNA. In this system,when the 5′ UTR ofpsbDis fused to the gene of interest,and theNac2gene is fused to an inducible nuclear promoter,the expression of target genes will be controlled in a Nac2-dependent manner (Ramundoet al.2013;Ramundo and Rochaix 2015;Rochaixet al.2021). When theNac2gene is fused to the promoter of cytochrome c6 (Cyc 6) gene,the target gene will be obviously repressed with the presence of copper in the growth medium and strongly expressed under copper deprivation or in the presence of nickel (Merchant and Bogorad 1987;Quinnet al.2002). Alternatively,when vitamin-repressible MetE promoter and Thi4 riboswitch are fused to theNac2gene,the target gene can be inactivated in a reversible way by supplying vitamin B12 and thiamine to the growth medium,respectively. This vitamin repressible system has been successfully used to investigate the roles of several essential chloroplast genes,includingrpoA,rps12,andclpP(Ramundoet al.2013,2014);additionally,it has shown great potential in expressing foreign proteins of commercial interests.Nevertheless,it is toxic and can severely affect cell growth.Using this inducible chloroplast gene expression system may induce the expression of the protein of interest once the cell culture reaches the appropriate cell density,minimizing the toxicity effect (Surzyckiet al.2009).
Selection of 3′ UTRThe 3′ UTRs of chloroplast mRNAs mainly perform the stabilizing function of mRNAs (Adams and Stern 1990). They are folded into stable stem-loop RNA secondary structures by inverted repeat (IR) base sequences with weak terminators,thereby protecting mRNA from 3′ to 5′ exonuclease degradation (Mondeet al.2000). Previous research has shown that the AUrich region of the 3′ UTR can enhance or inhibit translation when combined with different trans-acting factors (Kruyset al.1989;Kontoyianniset al.1999). This also leads to the speculation that it is not acis-element,but a specifictrans-acting factor,that determines the function of a specific 3′ UTRcis-element. Research on the expression of different 3′ UTRs with the same reporter gene has revealed that the RNA accumulation level may be affected by 3′UTR selection,particularly the stability of transcripts and 3′end processing (Rottet al.1998;Eiblet al.1999;Barneset al.2005;Tangphatsornruanget al.2011). For example,under the control of thepsbApromoter,the reporter GFP gene was combined with the 3′ UTR of several tobacco chloroplast genes and the terminator of theE.colirrnBoperon. The results showed that therrnBterminator resulted in the highest GFP transcript,followed bypsbAandpetD,and thenrbcL(Tangphatsornruanget al.2011).
Furthermore,it has been demonstrated that different combinations of 5′ UTR and 3′ UTR would result in different mRNA stability,which follows the order ofrbcLrbcL>rbcL-psbA>psbA-rbcL>psbA-psbA(5′ UTR-3′UTR),and the difference betweenrbcL-rbcLandpsbApsbAis approximately 3-fold (Eiblet al.1999).
Selection of fusion adaptorsIt remains a great challenge to express very small polypeptides in chloroplast due to the presence of specific proteases in plastids that can degrade polypeptides smaller than 65 amino acids. Hoelscheret al.(2022) have solved this problem by constructing protein fusions with an increase in the polypeptide size above the critical threshold for protease recognition,such as fusion of multiple AMPs separated by flexible linkers and/or fusion to SUMO(small ubiquitin-like modifier). Previous data have suggested that fusion to SUMO can provide a valuable tool for the efficient production and purification of recombinant protein from transplastomic plants,as it can improve protein accumulation presumably by increasing protein stability,enhance the solubility of AMPs,and thus facilitate the purification and avoiding the use of detergents for AMP extraction. Additionally,fusion to SUMO also alleviates the mutant phenotypes associated with fAMP expression.
3.4.Selection markers
Improving the selection efficiency for chloroplast transformation is one of the challenges to expanding the range of species for chloroplast transformation. There are numerous selection markers with various selection mechanisms (Yuet al.2020),and a commonly used selection marker in chloroplast transformation is the aadA cassette,which is characterized by spectinomycin/streptomycin antibiotic resistance properties.
As a widely used model plant,Arabidopsis thalianahas long suffered from lower chloroplast transformation efficiency (Sikdaret al.1998). One of the major causes is the low selection efficiency of spectinomycin in most Arabidopsis accessions. Recent research has revealed that when chloroplast translation is blocked by spectinomycin,the chloroplast-encodedaccDgene does not produce heterologous ACCase,but homomeric ACCase encoded by the nuclear geneAAC2can promote fatty acid biosynthesis,reducing the effect of lack of heteromeric ACCase in the process of tissue culture.Knockout of the Arabidopsis nuclear geneAAC2by CRISPR/Cas can improve the chloroplast transformation efficiency by about 100 folds (Yuet al.2017). Furthermore,the microcallus induced from roots can ensure the fertility in Arabidopsis (Rufet al.2019). Recently,knockout of theACC2gene inBrassica napuswas also found to result in the acquisition of a spectinomycin allergic phenotype,providing a useful resource for further improvement of chloroplast transformation inB.napus(LaMannaet al.2022). These findings provide important implications for high-efficiency chloroplast transformation in those crops with heteromeric and homomeric ACCases in their chloroplasts. However,these methods may be ineffective in most Poaceae species,including wheat and rice,since the heteromeric ACCase is replaced by a chloroplasttargeting homopolymerase encoded by nuclear genes(Huanget al.2002).
Furthermore,it is critical to remove the selection marker genes so as to further improve the social acceptance of chloroplast genetic engineering. Fortunately,Cre recombinase can well remove the selection marker in chloroplast transformation (Pradhanet al.2016;Tungsuchat-Huang and Maliga 2021).
3.5.Chloroplast genetic transformation of linear fragments
Traditional chloroplast transformation requires the construction of transformation vectors in a circle superhelix plasmid form that can be multiplated inE.coli.On the one hand,it is particularly cumbersome for the construction of multigene vectors,though some new tools for efficient vector construction have emerged,such as Gateway cloning (Gottschamelet al.2013)and MoChlo (Occhialiniet al.2019). On the other hand,the construction will be difficult when the vector contains genes encoding proteins that are toxic toE.coli.Therefore,it is particularly important to seek a simple and low-cost transformation method. Renet al.(2022) found that multiple linear DNA fragments obtained by PCR can be precisely integrated into the chloroplast genome with the assistance of homologous sequences at their ends.Homologous sequences of 200 bp or longer are sufficient,and the transformation efficiency is at least comparable to that of common vectors (Renet al.2022). It is a cloningfree chloroplast transformation technology and a good choice for chloroplast transformation with a single or a small number of fragments. Nevertheless,it remains to be determined whether it is suitable for the construction of synthetic pathways of multiple genes,and it is important to balance the relationship between the length of homology arms and the efficiency of fragment insertion.
4.Conclusion and outlook
As an important branch of plant genetic engineering,chloroplast transformation technology has successfully integrated more than 120 genes into plant chloroplast genomes with high-efficiency expression. These genetic transformations are intended to improve the resistance of plants to insects,bacterial,fungal,and viral diseases,develop different types of herbicides,enhance abiotic stress tolerance,utilize cytoplasmic male sterility,phytoremediate toxic metals,develop vaccine antigens,biopharmaceuticals,and industrial enzymes,and synthesize biofuels. Although numerous new transformation methods and regulatory elements have been developed over the past three decades,chloroplasts can be reliably transformed only in a limited number of algal and plant species. As for monocot cereal crops,although homoplastomic rice has been reported (Wanget al.2018),it remains a challenge for other crops due to the lack of suitable selection reagent/marker gene,highefficiency shoot regeneration system,cis-elements for expressing transgenic genes in non-green chloroplasts,and difficulty in obtaining homoplastomic plants.
On the other hand,although the emergence of genome editing methods,such as TALENS and CRISPR/Cas system,has significantly facilitated genome editing,the double membrane of organelles in higher plants prevents the import of most nucleic acids,which impedes the successful application of CRISPR/Cas to the editing of higher plant organelles (Leslie 2018). Recently,the application of transcription activator-like (TAL) effectorbased tools has led to dramatic changes in base editing since the modular assembly of TALE repeats can ensure recognition of the target site in organelle genomes (Maliga 2021). RNA editing of plant chloroplast genomes is essential for plant chloroplast development and photosynthetic protein accumulation (Caoet al.2023;Chenet al.2023). Currently,many base editors have been successfully applied to plant organelle genome editing. Chloroplast-targeted (pt) cytosine base editor (ptpTALECD) (Nakazatoet al.2021) and DddAderived cytosine base editor (DdCBE) (Kanget al.2021) can specifically introduce homoplasmic C-to-T mutations into target windows in the chloroplast genome.Moreover,transcription activator-like (TAL) effectorlinked deaminases can induce heritable homogeneous A-to-G mutation in chloroplast DNA,leading to phenotypic changes (Moket al.2022). These methods are suitable to be applied to species based on nuclear transformation to modify chloroplast-encoded endogenous genes where the chloroplast transformation system has yet to be established.
A brief report in 2015 has validated that CRISPR/Cas can target specific sites in the mitochondrial genome(Joet al.2015),indicating that CRISPR/Cas may also be used in organelles. To overcome the difficulty in organelle genome editing,a novel “editing plasmid”was applied for the CRISPR editing of organelle genomes,which comprises a cassette for Cas9-type endonucleases,an expression guide RNA,donor DNA,and a cassette for a selection marker. The successful incorporation of donor DNA into theChlamydomonaschloroplast genome will only occur in the presence of Cas9/gRNA in the editing plasmid,while the frequency of substitution and INDEL mutation at a cleavage site was below detection,indicating that homology-directed DNA repair and replacement are the main outcomes of Cas9 induced cleavage in organelles (Yooet al.2020).More importantly,Yooet al.(2020) found no evidence of Cas9 toxicity in stably transformed algal organelles,unlike that previously reported for nuclear expression of Cas9 expression inChlamydomonas(Jianget al.2014).The approach is expected to pave the way for precisely editing organelle DNA and introducing new alleles and genes without leaving any trace of transgenes. However,challenges remain in increasing the efficiency of organelle genome editing and promoting homoplasmy.
This review has demonstrated the significant advantages of technical advances in chloroplast transformation. However,many challenges or obstacles exist to extending the chloroplast transformation from model species and cultivars to other species and cultivars.
Despite many obstacles,chloroplast genetic engineering has hugely benefited human society.Therefore,more efforts should be made to expand the application of chloroplast genetic engineering further.
Acknowledgements
This work was funded by the Foundation of Hubei Hongshan Laboratory,China (2022hszd014) and the National Natural Science Foundation of China (31771752).
Declaration of competing interest
The authors declare that they have no conflict of interest.
 Journal of Integrative Agriculture2023年7期
Journal of Integrative Agriculture2023年7期
- Journal of Integrative Agriculture的其它文章
- Understanding changes in volatile compounds and fatty acids of Jincheng orange peel oil at different growth stages using GC-MS
- Untargeted UHPLC-Q-Exactive-MS-based metabolomics reveals associations between pre-and post-cooked metabolites and the taste quality of geographical indication rice and regular rice
- A double-layer model for improving the estimation of wheat canopy nitrogen content from unmanned aerial vehicle multispectral imagery
- The potential of green manure to increase soil carbon sequestration and reduce the yield-scaled carbon footprint of rice production in southern China
- lmprovement of soil fertility and rice yield after long-term application of cow manure combined with inorganic fertilizers
- A novel short transcript isoform of chicken lRF7 negatively regulates interferon-β production
