Increased CD4/CD8 Lymphocyte ratio predicts favourable neoadjuvant treatment response in gastric cancer: A prospective pilot study
Daniel Skubleny, Andrea Lin, Saurabh Garg, Ross McLean, Michael McCall, Sunita Ghosh, Jennifer L Spratlin,Daniel Schiller, Gina Rayat
Daniel Skubleny, Department of Surgery, University of Alberta, Edmonton T6G 2R3, AB, Canada
Andrea Lin, Saurabh Garg, Michael McCall, Daniel Schiller, Gina Rayat, Department of Surgery, University of Alberta, Edmonton T6G 2R3, Alberta, Canada
Ross McLean, Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton T6G 2R3, Alberta, Canada
Sunita Ghosh, Department of Oncology, University of Alberta, Edmonton T6G 2R3, Alberta, Canada
Jennifer L Spratlin, Department of Oncology, Cross Cancer Institute, University of Alberta, Edmonton T5G 1Z2, AB, Canada
Abstract BACKGROUND Despite optimal neoadjuvant chemotherapy only 40% of gastric cancer tumours achieve complete or partial treatment response. In the absence of treatment response, neoadjuvant chemotherapy in gastric cancer contributes to adverse events without additional survival benefit compared to adjuvant treatment or surgery alone. Additional strategies and methods are required to optimize the allocation of existing treatment regimens such as FLOT chemotherapy (5-Fluorouracil, Leucovorin, Oxaliplatin and Docetaxel). Predictive biomarkers detected using immunohistochemistry (IHC) methods may provide useful data regarding treatment response.AIM To investigate the utility of CD4, CD8, Galectin-3 and E-cadherin in predicting neoadjuvant FLOT chemotherapy tumour response in gastric adenocarcinoma.METHODS Forty-three adult patients with gastric adenocarcinoma, of which 18 underwent neoadjuvant chemotherapy, were included in a prospective clinical cohort. Endoscopic biopsies were obtained from gastric cancer and normal adjacent gastric mucosa. Differences in expression of Galectin-3, Ecadherin, CD4+ and CD8+ molecules between tumours with and without treatment response to neoadjuvant chemotherapy were assessed with IHC. Treatment response was graded by clinical pathologists using the Tumour Regression Score according to the College of American Pathologists criteria. Treatment response was defined as complete or near complete tumour response, whereas partial or poor/no response was defined as incomplete. Digital IHC images were annotated and quantitatively assessed using QuPath 0.3.1. Biomarker expression between responsive and incomplete response tumours was assessed using a two-sided Wilcoxon test. Biomarker expression was also compared between normal and cancer tissue and between 15 paired tumour samples before and after chemotherapy. We performed a preliminary multivariate analysis and power analysis to guide future study. Statistical analyses were completed using R 4.1.2.RESULTS The ratio between CD4+ and CD8+ lymphocytes was significantly greater in treatment responsive tumours (Wilcoxon, P = 0.03). In univariate models, CD4+/CD8+ ratio was the only biomarker that significantly predicted favourable treatment response (Accuracy 86%, P < 0.001). Using a glmnet multivariate model, high CD4+/CD8+ ratio and low Galectin-3 expression were the most influential variables in predicting a favourable treatment response. Analyses of paired samples found that FLOT chemotherapy also results in increased expression of CD4+ and CD8+ tumour infiltrating lymphocytes (Paired Wilcoxon, P = 0.002 and P = 0.008, respectively). Our power analysis suggests future study requires at least 35 patients in each treatment response group for CD8 and Galectin-3 molecules, whereas 80 patients in each treatment response group are required to assess CD4 and E-cadherin biomarkers.CONCLUSION We demonstrate that an elevated CD4+/CD8+ Ratio is a promising IHC-based biomarker to predict favourable treatment response to FLOT neoadjuvant chemotherapy in locally advanced gastric cancer.
Key Words: CD4; CD8; Galectin-3; Neoadjuvant chemotherapy; Treatment response; Gastric cancer
INTRODUCTION
Gastric cancer is the fifth most common cancer and the third most common cause of cancer death worldwide[1-3]. The poor prognosis associated with gastric cancer is in part related to significant tumour molecular heterogeneity[4-6]. Despite insight gained from extensive genomic and transcriptomic profiling, molecular classification systems such as those proposed by The Cancer Genome Atlas and Asian Cancer Research Group have yet to manifest improvement in the clinical management of gastric cancer.
In North America, the standard of care for locally advanced gastric cancer is neoadjuvant chemotherapy with 5-fluorouracil, leucovorin, oxaliplatin and docetaxel (FLOT4)[7]. Advantages of neoadjuvant chemotherapy include improved survival compared to surgery alone, greater R0 resection and reduction in nodal stage[8-11]. Previous research has demonstrated that pathologic complete response (pCR) or treatment response defined as Tumour Regression Grade 1-3 is significantly associated with improved prognosis[12,13]. However, pCR occurs in only 3-15% of cases and complete or partial response in approximately 40% of patients[9,12,14]. Irrespective of efficacy, cytotoxicity of chemotherapy is associated with adverse events including peripheral neuropathy, neutropenia, infection or death[7]. Prior evidence suggests that, in the absence of treatment response, neoadjuvant chemotherapy in gastric cancer contributes to adverse events without additional benefit compared to adjuvant treatment or surgery alone[9,15]. Specific treatment response may be related to underlying tumour biology as exposure to neoadjuvant chemotherapy in microsatellite instability in gastric cancer has been demonstrated to relate to worse survival outcomes[16-18]. Thus, in order to improve outcomes, it is of paramount importance to identify clinicopathologic or molecular biomarkers to identify treatment responders.
Immunohistochemistry (IHC) is a proven molecular pathology technique with a record of providing prognostic and therapeutic biomarkers in oncology. In gastric cancer, prominent IHC-based biomarkers may be prognostic or therapeutic as in the case of E-cadherin and human epidermal growth factor receptor 2, respectively[19,20]. However, there is a lack of predictive biomarkers to inform treatment response to more common regimens such as neoadjuvant chemotherapy.
Here we investigate a panel of biomarkers that we hypothesize may provide value in predicting tumour response. Galectin-3 is a lectin protein that facilitates cancer tumorigenesis and prognosis[21-24]. Pre-clinical models suggest that increased Galectin-3 expression is associated with chemotherapy resistance[25,26]. Recent work has implicated cell-surface expression of Galectin-3 with chemoresistance in gastrointestinal cancer stem cells[27]. E-cadherin is a cell-cell adhesion molecule that plays an important role in gastric cancer development, classification and prognosis[4,5,28]. In-vitro study has previously suggested that germline mutations in E-cadherin related to Hereditary Diffuse Gastric Cancer increases chemoresistance to taxol based agents[29]. However, study of breast cancer cell lines have identified heterogenous effects of E-cadherin expression on chemotherapy response[30,31]. We also assess whether CD4+and CD8+tumour infiltrating lymphocytes (TILs) and the relative proportion of these cells influence neoadjuvant chemotherapy response. The CD4/CD8 ratio is a marker of immune effector function and is associated with multiple disease states. A normal circulating CD4/CD8 ratio ranges from 1.5-2.5, and lower ratios in resident tissues or circulation are related to worse HIV related outcomes, cardiovascular disease and cancer[32]. Both CD4+and CD8+T cells are essential components to the tumour microenvironment and their composition in relationship to other immune cells such as macrophages, antigen presenting cells and natural killer cells influence the effectiveness of the host response to cancer[33]. Increasing evidence recognises the association of greater TILs to favourable cancer prognosis and chemotherapy response in colon and gastric cancer[34-39]. To date, no studies have investigated the role CD4+or CD8+TILs in neoadjuvant chemotherapy response for gastric cancer.
To guide future studies, we performed a prospective pilot study to evaluate if these selected biomarkers provide predictive value in evaluating treatment response following neoadjuvant FLOT chemotherapy.
MATERIALS AND METHODS
Study design
We performed this single-center, prospective pilot study at the University of Alberta in Edmonton, Alberta, Canada from January 2018 to January 2022. All human clinical participants consented according to the approved ethics protocol granted by the Health Research Ethics Board of Alberta (Study ID: HREBA.CC-17-0228_REN5). Treatment na?ve Stage I-IV sporadic gastric adenocarcinoma patients aged greater than 18 years were included. A subset of patients enrolled was allocated to a second cohort on the basis of receiving curative intent neoadjuvant FLOT chemotherapy (Figure 1). Patients with a known inherited oncogenic germline mutation or hereditary syndrome (i.e.,Familial Adenomatous Polyposis) were excluded.
Specimens were retrievedviaendoscopic biopsy at the time of diagnosis, screening laparoscopy or at the time of surgical resection at the Walter C Mackenzie Health Sciences Centre or Royal Alexandra Hospital. Normal biopsies were obtained from gastric mucosa greater than 5 cm away from the cancerous lesion or associated gastritis. The initial study protocol retrieved two tissue biopsies for permanent pathology, however, following interim review four biopsies were retrieved thereafter. The presence of cancer in specimens was confirmed by a gastrointestinal pathologist. In the absence of cancer, clinical formalin-fixed paraffin-embedded pathology blocks were retrieved when available. In clinical samples with treatment effect, residual cancer cells were detected using anti-pan cytokeratin (Abcam, clone C-11, ab7753) IHC staining followed by the manual assembly of tissue microarray (TMA) blocks with 4mm cores of regions containing residual tumour.
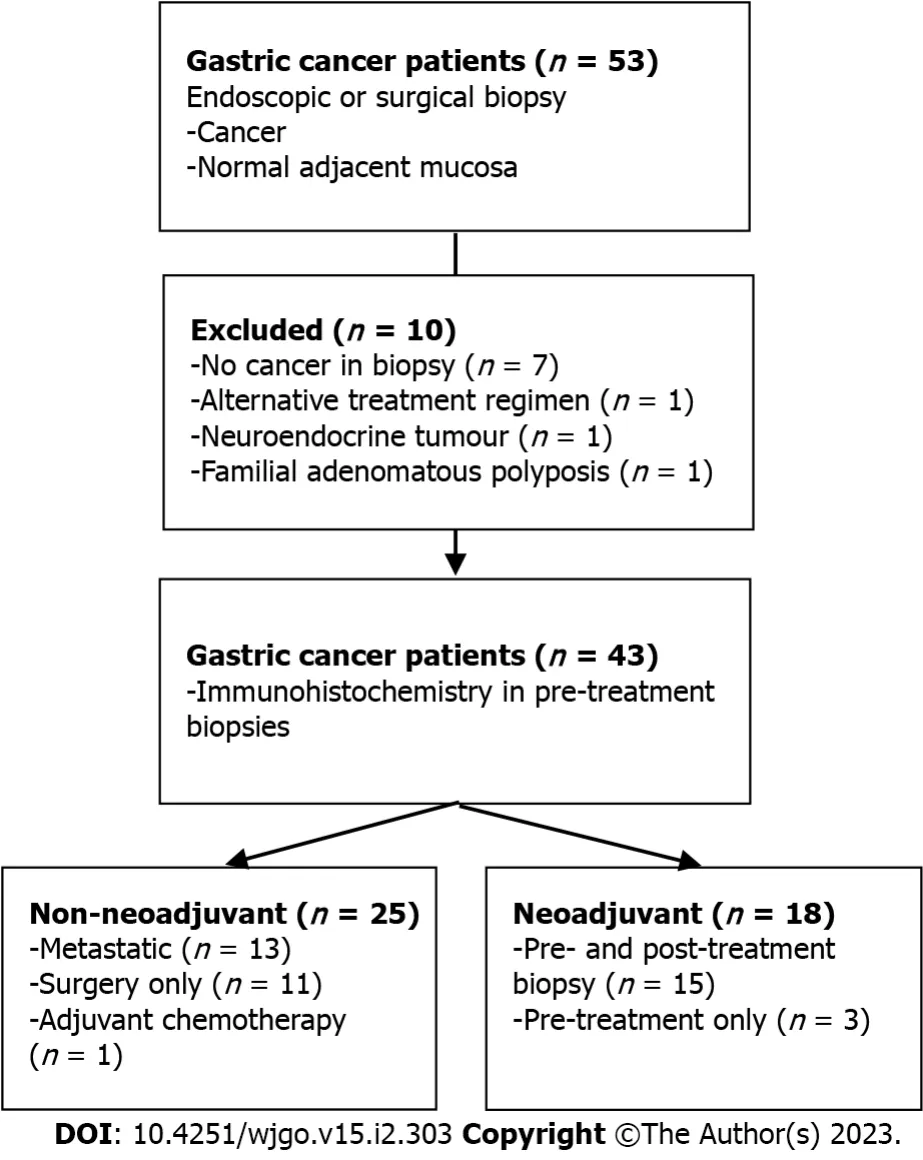
Figure 1 Study overview. Flow chart outlining study design and patient allocation.
Our primary outcome for all patients was the difference in expression of selected biomarkers between normal and cancer tissue. In the subgroup of patients receiving neoadjuvant chemotherapy, our primary outcome was the difference in expression between tumour treatment response and incomplete treatment response. We also evaluated the difference in expression of biomarkers in paired samples before and after chemotherapy treatment.
Treatment response was retrieved from clinical pathology reports. The Tumour Regression Score was graded according to the College of American Pathologists and National Comprehensive Cancer Network protocol on a 4-point scale (0 = Complete response, 1 = near complete response, 2 = partial response, 3 = poor or no response)[40]. In accordance with prior studies, treatment response was expressed as a binary variable consisting of response and incomplete response categories[12]. Responsive tumours included complete and near-complete responses, whereas incomplete responses included partial, and poor no response. Patients who progressed to metastasis while receiving neoadjuvant treatment were classified as an incomplete response.
IHC
Tissue specimens of normal and cancer tissue were fixed in zinc-formalin (Z-Fixx, Sigma-Aldrich) for 24 h, washed three times and stored in 70% ethanol prior to preservation in paraffin. Briefly, 4 μm tissue sections were deparaffinized in Histoclear (National Diagnostics) and rehydrated. Endogenous peroxidases were quenched using 3% hydrogen peroxide in methanol for 5 min. Microwave heat induced epitope retrieval was performed using Sodium Citrate (pH 6, heated to 94 degrees Celsius in 1-min intervals followed by 9 min continuous heat) for E-cadherin and Tris-ethylenediaminetetraacetic acid (pH 9, heated to 94 degrees Celsius in 1-min intervals followed by 8 min 30 s continuous heat) for CD4 and CD8. Non-specific staining was blocked using 20% normal goat serum (Jackson Laboratories) for E-cadherin, CD4 and CD8 or 2% Fetal Bovine Serum (Gibco) in 1X phosphate buffered saline for Galectin-3 for 20 min followed by avidin and biotin blocking (Vector Laboratory, SP-2001)permanufacturer’s protocol. Tissue sections were stained with primary antibodies anti-E-cadherin (1:25, 1.5 h room temperature, ThermoFisher Scientific, clone 4A2C7, 33-4000), anti-Galectin-3 (1:200, 30 min room temperature, Cedarlane, clone M3/38, CL8942AP), anti-CD4 (1:200, overnight at 4 degrees, Abcam, clone EPR6855, ab133616) or anti-CD8 alpha (1:200, overnight at 4 degrees Celsius, Abcam, ab4055). All biotinylated immunoglobulin G secondary antibodies were incubated at 1:200 for 30 min at room temperature, including rabbit anti-rat for Galectin-3 (Vector Laboratories, BA-4001), goat-anti-rabbit for CD4 (Vector Laboratories, BA-1000) and goat-anti-mouse for E-cadherin and CD8 (Jackson ImmunoResearch, 115-065-003). Antibody detection was performed using avidin-biotin complex/horseradish peroxidase (Vector Laboratories) and 3,3-diaminobenzidine tetrahydrochloride (DAB, Abcam, ab64238)permanufacturer’s protocol. Stained tissue sections for E-Cadherin, CD4 and CD8 were counterstained with Harris’ hematoxylin (Fisher Scientific) and Harris’ hematoxylin and eosin (Fisher Scientific) for Galectin-3.
Histology imaging and quantification
Histology images were captured at 20 times magnification using a Leica Aperio CS2 digital slide scanner. Digital pathology quantification of antibody expression was performed using QuPath version 0.3.1 (Figure 2A)[41]. Briefly, digital images were uploaded and the tumour and immediate tumour-host interface were annotated as a single region of interest. Stain vectors were estimated using default settings for each sample. For CD4, CD8 and Galectin-3, positive cells were detected using default nucleus DAB optical density settings. The CD4/CD8 ratio was calculated as the proportion of positively stained CD4 cells divided by the proportion of positively stained CD8 cells. For E-cadherin, both the proportion of positive cells and H-score was calculated. Annotated cell regions were assessed for accuracy and in the event of background or non-specific staining positive cell threshold values were adjusted to reflect true positive staining. The H-score provides a consensus scoring method for evaluating immunostaining across a gradient of intensity (Equation 1). As defined in McClellandet al[42], H, M and L denotes high, medium and low intensity staining. Cells without staining are denoted N for negative staining.
Statistical analysis
Statistical analyses were completed using R version 4.1.2[43]. The statistical methods of this study were reviewed by Dr. Ghosh and Dr. Skubleny from the University of Alberta. Differences between groups were assessed with a Wilcoxon two-sample test for independent samples and two-tailed paired Wilcoxon test for paired samples. Statistical significance was defined at alpha = 0.05. Multiple comparisons corrections were not made for our main outcomes given our prespecified analyses, but the possibility of false positive results is noted. Summary of continuous variables is expressed as median with interquartile range. Categorical variables are expressed as absolute number of cases and percent proportions.
The ability of biomarkers to predict treatment response was assessed using the caret package in R[44]. Briefly, out-of-sample resampling accuracy was estimated for each biomarker as well as the combination of all biomarkers using 1000 bootstraps with replacement. Continuous variables were centered and scaled. Logistic regression models were used for single biomarker estimates and a regularized ElasticNet model implemented in glmnet was used for estimates containing all biomarkers[45]. Model significance was tested using a one-sided binomial test comparing the estimated model accuracy to the No Information Rate (NIR). The NIR is defined as the largest proportion of observed classes, or the maximum accuracy of a classifier if it predicted the majority class every time.
Sample size calculations were performed using the MKpower package in R. Two-sample Wilcoxon distributions were generated using the mean and standard deviation from our pilot study sample. The normality of the distribution for each biomarker’s expression levels were confirmed with a Shapiro-Wilk test. Random sampling from a truncated normal distribution constrained between 0 and 100 was performed for a series of samples sizes ranging from 10 to 120, in intervals of 10. The empirical power (beta) for each sample size was calculated using Monte-Carlo simulations with 1000 iterations for a specified type-I error rate (alpha = 0.05).
RESULTS
Patient demographics
Fifty-three patients were consented for this pilot study. Ten patients were excluded: One patient was diagnosed with Familial Adenomatous Polyposis, one was found to have neuroendocrine tumour pathology, one gastroesophageal junction tumour received alternative neoadjuvant therapy and seven patients were excluded due to inadequate tissue biopsies. Of note, an interim analysis of our protocol after enrolling the first 20 patients determined a biopsy accuracy rate of 60% for treatment na?ve specimens and 25% for biopsy following neoadjuvant treatment. This prompted a change in study protocol to retrieve 4-8 tissue biopsiespersample.
A total of 43 patients were available for analysis, of which 18 (42%) underwent neoadjuvant chemotherapy during our study period. Baseline demographics are included in Table 1. Median age was 65 (60, 75) and the majority of patients were male (70%). Tumour pathology was represented by all TNM stages but a preponderance of high grade (72%), proximal stomach (60%) and diffuse type (63%) tumours were present.H. pyloristatus was available for 32 patients, of which the majority were negative (69%) and one was previously treated. Total gastrectomy was performed in nearly half of all patients and comprised 59% of all surgical resections.
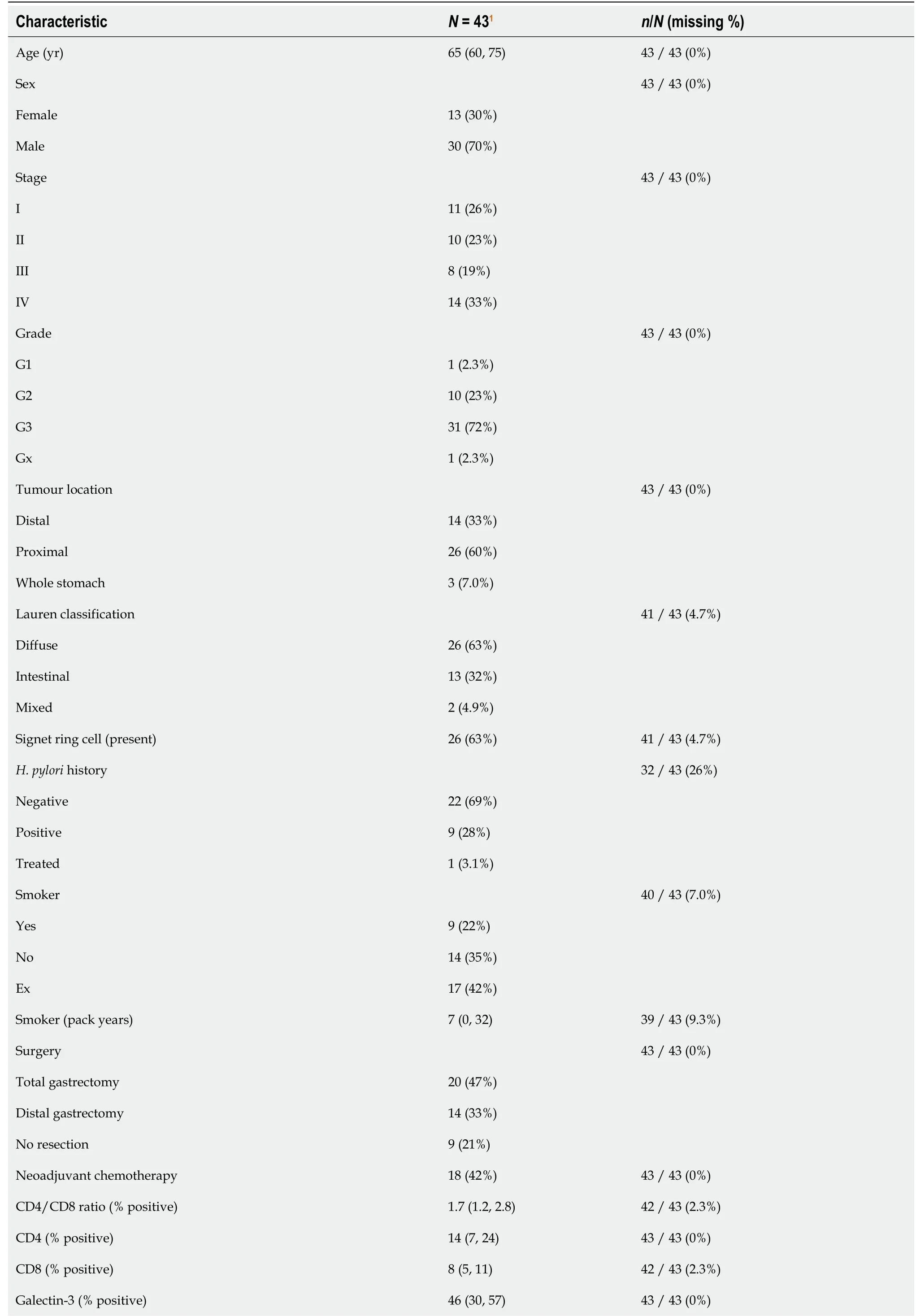
Table 1 Baseline demographics

1Median (IQR); n (%).
Expression of biomarkers in normal and cancer tissues
Representative images of each IHC stain within the 75thand 25thpercentile of expression is presented in Figure 2B. Staining for E-cadherin was only identified on cell membranes of gastric epithelium. Galectin-3 exhibited heterogeneous staining and was identified in nuclei, cytoplasm, and surrounding tumour stroma. The presence of Galectin-3 was often sporadic with distinct regions representing intense positive stain followed by fairly abrupt transition to moderate positivity. CD4 and CD8 positive staining was identified on the cell membrane of lymphocytes.

Figure 2 Immunohistochemistry stains and expression of biomarkers in treatment na?ve normal and cancer tissue. A: Representative images of QuPath digital pathology annotation using CD4 immunohistochemistry (IHC) at 10X magnification. Raw images (left) are processed and regions of interest are identified according to our methods. The annotated image (right) demonstrates the calculation of positive stained cells (red) and negative cells (blue); B: Representative IHC images taken at 10X magnification for each respective biomarker identified on the y-axis. Images of expression values within the 75th and 25th percentile are presented in the left and right columns, respectively. Arrows demonstrate positive staining in low expression specimens; C: Boxplot comparison of expression for each respective biomarker in treatment naive normal and cancer tissue. The IHC biomarker is labeled on the heading of each graph. The y-axis represents IHC score, which is the percent of positive stained cells for Galectin-3, CD4, CD8 and E-cadherin and the H-score for E-cadherin H-score plot. The x-axis labels the distribution corresponding to normal (blue) and cancer (red) tissue. The raw P value for Wilcoxon tests is annotated in each panel.
Galectin-3 was the most abundant molecule with a median expression of 46% (30, 57), followed by Ecadherin, CD4 and CD8 (Table 1). The E-cadherin H-score (median 22 (7, 40)) closely approximated the proportion of E-cadherin positive cells (median 18 (6, 28)). Greater H-score values in the upper quartile reflected the presence of high staining intensity in positive cells.
Significantly increased expression of CD4, Galectin-3 and CD4/CD8 Ratio was identified in cancer tissue relative to normal adjacent tissue controls (Wilcoxon,P= 0.035,P= 0.020 andP= 0.018 respectively) (Figure 2C). The distribution of IHC scores between normal and cancer tissue for CD4 and Galectin-3 was relatively uniform, whereas differences in CD4/CD8 Ratios were dominated by sample outliers with large cancer IHC scores. In agreement with historical study, E-cadherin positivity and Hscore was significant decreased in cancer tissue relative to normal. (Wilcoxon,P< 0.0001 andP< 0.001, respectively).
There were no statistically significant associations between relevant clinicopathologic factors and the expression of any biomarker for stage, lymphovascular invasion, perineural invasion, carcinomatosis, tumour grade or location (Supplementary material). The proportion of E-cadherin positive cells was significantly different according to Lauren Class, with relatively fewer positive cells present in diffuse and mixed type cancers (Kruskal-Wallis,P= 0.043).
Association of biomarker expression with exposure to neoadjuvant chemotherapy
We compared the expression of biomarkers in 15 paired tumour samples from the same patient before and after neoadjuvant FLOT to evaluate the effect of treatment on biomarker expression. All pretreatment specimens were obtained by endoscopic biopsy and thus were restricted mainly to the mucosa and lamina propria. The majority of post-treatment samples were analyzed as TMA cores from surgical resection specimens (TMA cores = 87%vsbiopsy = 13%) in which residual tumour was present in mucosa, submucosa and muscularis.
We found significantly increased association of tumour cells with CD4+and CD8+TILs following neoadjuvant chemotherapy (Paired Wilcoxon,P= 0.002 andP= 0.008, respectively) (Figure 3A). In contrast, E-cadherin positivity and H-score significantly decreased in post-treatment samples (Paired Wilcoxon,P= 0.035 andP= 0.04, respectively). This was likely in part due to differences in tumour cell depth of invasion between pre-treatment biopsy and post-treatment TMA cores. CD4/CD8 Ratio expression remained relatively stable within samples except for one patient (Figure 3A).
High CD4/CD8 ratio is associated with treatment response
Figure 3B outlines the relationship of biomarker expression to treatment response between pre- and post-treatment cancer specimens. For all analyses, we observed incomplete response in 14 patients (Partial = 9, Poor or No = 4, Progression to metastasis = 1) and response in 4 patients (Complete = 1, Near Complete = 3). Statistically greater CD4/CD8 Ratios were observed in pre-treatment cancer biopsies compared to incomplete responders (Wilcoxon,P= 0.025). Clinicopathologic characteristics were similar between treatment response groups (Table 2).
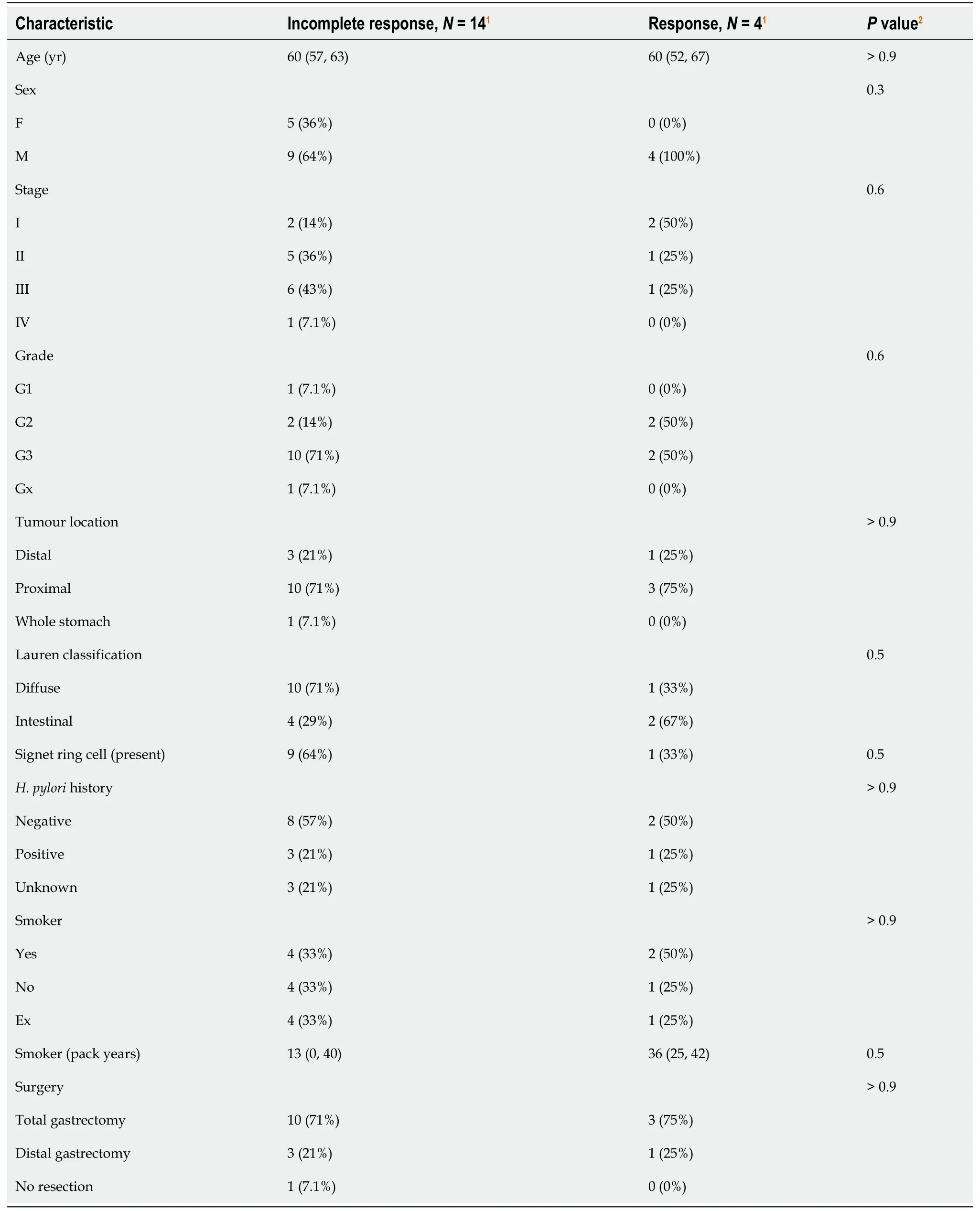
Table 2 Clinicopathologic factors according to treatment response
Next, we explored the utility of individual biomarkers (Models 1-6) and the combination of all biomarkers (Model 7) in predicting treatment response scores (Figure 3C). Given the small sample size and eventspervariable, we used out-of-sample estimates from 1000 bootstraps to limit bias by favouring pessimistic estimates of model accuracy. In this dataset, all biomarkers were effective at predicting incomplete tumour response (Sensitivity range 88-98%) but suffered from poor specificity (range 0-44%). CD4/CD8 Ratio was the only variable that provided significant model performance (Accuracy > NIR, one-sided binomial,P< 0.001). The ElasticNet model using CD4/CD8 Ratio, CD4, CD8, Galectin-3 and ECAD H-score as independent variables provided a mean accuracy greater than the NIR but failed to achieve statistical significance (P= 0.26).
The optimal glmnet model provided coefficients for all variables despite tuning parameters allowing for L2 regularization (alpha = 0). To guide future studies, we evaluated the contribution of all biomarker variables to the predictive model using the final regularized ElasticNet coefficients (Figure 3D). The absolute value of coefficients found CD4 /CD8 Ratio and Galectin-3 to provide the greatest influence in predicting favourable tumour response. Specifically, tumour response was associated with increasing CD4/CD8 Ratio and decreasing Galectin-3, respectively.
Sample size calculations
To inform future studies we performed sample size calculations using our pilot study sample distributions. In particular, we were interested in identifying the sample sizes required to evaluate the utility of biomarkers in explaining tumour response using a two-sample Wilcoxon test. In Figure 3E, we observe that CD8 and Galectin-3 require similar sample sizes of 30 and ~35 in each treatment response group to achieve adequate power. The relationship between sample size and empirical power was nearly identical for CD4 and E-cadherin, which were calculated to require ~70 and 80 samples in each group, respectively.
DISCUSSION
In this pilot study, we present the utility of IHC-based expression of Galectin-3, E-cadherin, CD4 and CD8 in predicting treatment response to the neoadjuvant chemotherapy regimen FLOT4. First, we establish that Galectin-3, CD4, E-cadherin and the CD4/CD8 Ratio expression are significantly different between cancer and normal adjacent tissue. These findings suggest that these markers are intrinsic to the tumour or tumour microenvironment and thus may provide prognostic or predictive yield. Next, we establish that the CD4/CD8 Ratio is significantly greater in tumours with complete or partial response to neoadjuvant chemotherapy. In preliminary univariate and multivariate machine learning models, the CD4/CD8 Ratio was the only significant predictive marker of treatment response with an accuracy of 86%. Finally, we demonstrate that the tumour-specific expression of CD4, CD8 and E-cadherin is significantly modified in paired tumour samples before and after chemotherapy.
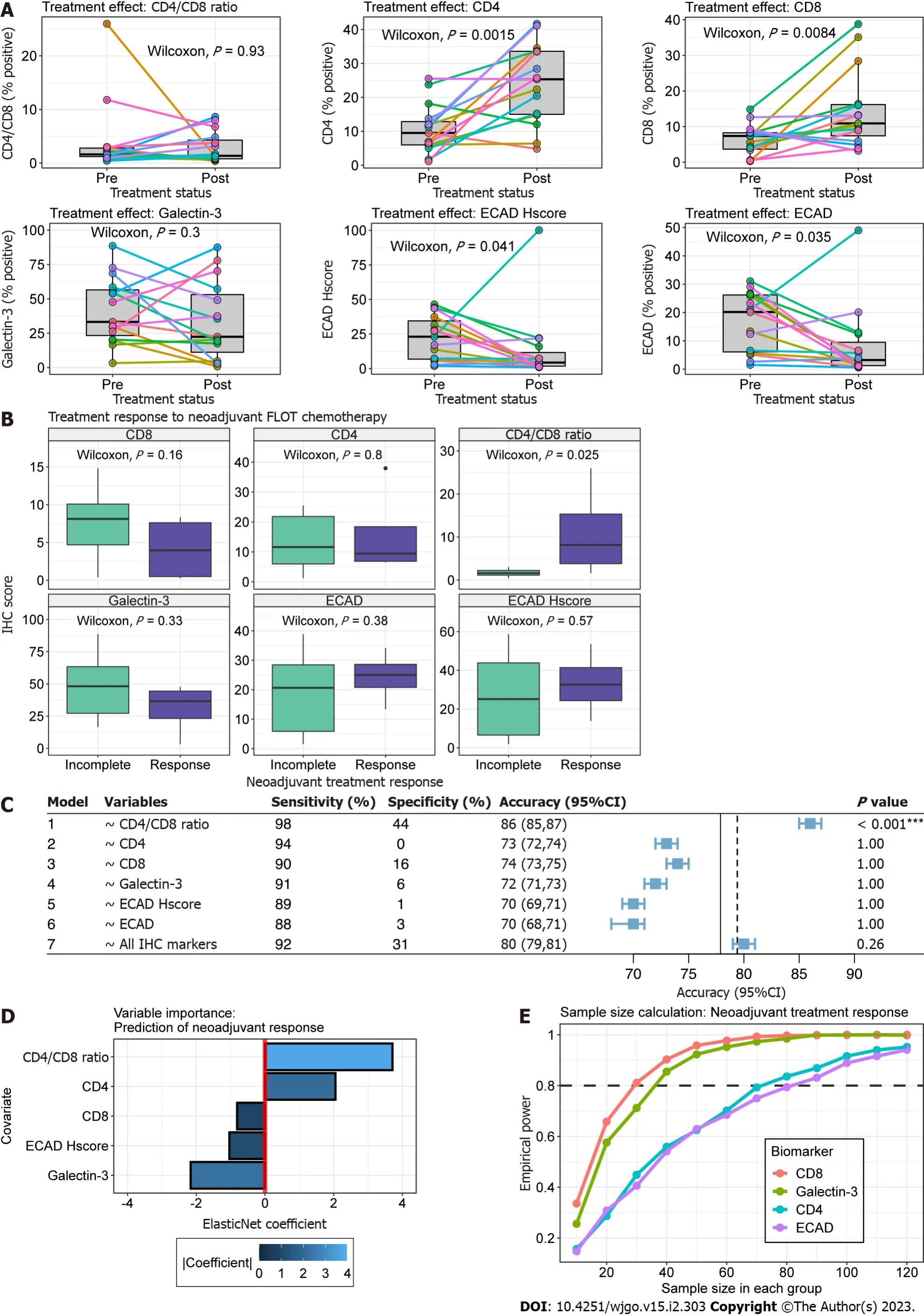
Figure 3 Association between biomarker expression and neoadjuvant 5-fluorouracil, leucovorin, oxaliplatin and docetaxel chemotherapy.A: Paired boxplots for biomarker expression pre- and post-neoadjuvant chemotherapy. Each coloured point and line correspond to a single patient. Boxplots in grey represent the distribution of expression for all patients before and after chemotherapy. Paired Wilcoxon P value is present in each plot; B: Boxplot comparison of expression for each respective biomarker between treatment response (purple) and incomplete response (green). The immunohistochemistry (IHC) biomarker is labeled on the heading of each graph. The y-axis represents IHC score, which is the percent of positive stained cells for Galectin-3, CD4, CD8 and E-cadherin and the H-score for ECAD H-score plot; C: Forest plot for metrics of ElasticNet models. Models were constructed using the treatment response as the dependent variable and the corresponding independent variable(s) identified in the variable column. The plot represents the out-of-sample accuracy (blue square) and 95% confidence intervals (whiskers) for models estimated from 1000 bootstraps with replacement. The no information rate, defined as the maximum accuracy of a classifier if it predicted the majority class every time, is shown by the solid and dotted vertical lines for univariable and multivariable models, respectively; D: Barplot of model coefficients from multivariable glmnet model. The y-axis represents model covariates and the x-axis the coefficient value. Treatment response is related to increasing covariate values or decreasing covariate values for positive and negative coefficients, respectively. The absolute importance of the coefficient is shown in blue according to the scale legend; E: Lineplot illustrating monte-carlo simulations for two-sample Wilcoxon sample size calculations. The y-axis is the empirical power and the x-axis is the sample size in each group. Each coloured line corresponds to a biomarker labelled according to the legend.
Several potentially useful approaches for determining treatment response have previously been recognized. Clinical or pathologic factors including age, tumour grade, signet cell pathology, serum carcinoembryonic antigen, various circulating lymphocyte populations and tumour size are significant predictors of tumour response[46-48]. The majority of predictive tumour biomarker research in gastric cancer has focused on identifying molecules associated with adjuvant chemotherapy response. For example, a multivariable model utilizing the measurement of several TIL populations in 879 patients provided 3-year survival prediction accuracies of 79 and 84% for surgery alone and adjuvant chemotherapy populations, respectively[34]. In the neoadjuvant setting, a post-hoc analysis of 83 patients in the COMPASS trial identified several candidate gene expression based-biomarkers such asTIMP1andDSG2using quantitative real-time polymerase chain reaction[49]. Other studies to identify treatment response have used microRNAs, exosomes, inflammatory markers or medical imaging data[50]. Although predictive and prognostic factors identified in these studies show promise, there is limited external validity of these studies and clinical implementation is yet to be achieved.
This is the first study to evaluate the role of tumour-associated CD4/CD8 Ratio in gastric cancer neoadjuvant chemotherapy response. Increasing evidence has demonstrated the coordinated role of CD4+and CD8+T-cells in mediating tumour immune surveillance, immunotherapy response and cancer prognosis[51]. Sustained and effective tumour immune response requires CD4+T-cells, which potentiate effector CD8+responseviasecretion of cytokines such as interleukin-2, participate in direct anti-tumour effectsviainterferon-gamma and tumour necrosis factor, or facilitate antibody mediated humoral response from B-cellsviaCD40 Ligand binding[52]. Indeed, research evaluating chimeric antigen receptor (CAR) T-cell immune populations demonstrate increased anti-tumour activity with increasing CD4/CD8 ratio[53]. Yanget al[54] also demonstrated that CD4+CAR T cells are more effective at maintaining anti-tumour activityin vivocompared to CD8+ CAR T cells that are prone to exhaustion and apoptosis. Furthermore, in native tumour microenvironments increasing CD4/CD8 Ratio of the tumour-host interface in triple negative breast cancer is associated with improved overall and recurrence-free survival[51].
The dynamic increase in TIL expression following neoadjuvant chemotherapy in our pilot study also replicates previous findings. Significant work in breast cancer has implicated the pattern of TIL changes following chemotherapy to treatment response. In particular, greater CD4+T-cell expression is associated with pathologic complete response[55]. Also, decreased immune infiltration is a notable characteristic of residual tumours following neoadjuvant chemotherapy relative to pre-treatment biopsy[55]. Continued evaluation of the relationship of dynamic changes in CD4 and CD8 populations in gastric cancer are required to fully leverage these biomarkers.
Our study design is intended to provide a reproducible and externally valid method of biomarker analysis. Using IHC allows for easier clinical implementation given that common pathology workflows already include IHC analysis. Our use of open-source digital pathology software such as QuPath also provides a standardized basis to internally and externally validate our method in future studies. Digital pathology allows annotation and measurement of regions of interest within the software and thus eliminates the need for complex physical microdissection utilized in other biomarker studies.
The main limitation of this study is the low enrollment of curative intent patients. This is likely due to low disease incidence in our population but also may be related to the severe acute respiratory syndrome coronavirus 2 pandemic. Given our rate of patient enrollment, future study should prioritize increasing sample size by using a retrospective design in order to provide more accurate estimates for future multi-centre prospective study. Our sample size calculation suggests that a limited retrospective study with approximately eighty-five patients in each group will provide adequate power to assess these relationships.
CONCLUSION
The CD4/CD8 Ratio is a promising IHC-based biomarker with therapeutic implications for response to neoadjuvant chemotherapy in locally advanced gastric cancer. Future inquiry should focus on evaluating the prognostic value of these markers and the generation of a sufficient sample size to establish a predictive model for potential future clinical use.
ARTICLE HIGHLIGHTS
Research background
Neoadjuvant chemotherapy for gastric cancer is standard of care in western nations. Despite optimal therapy, only 40% of patients achieve complete or near complete treatment response. Treatment response following neoadjuvant chemotherapy is associated with overall survival. Thus, it is of critical importance to identify biomarkers capable of predicting which patients will achieve a favourable response to neoadjuvant chemotherapy in order to optimize survival outcomes.
Research motivation
Personalized medicine is predicated on providing the right treatment for the right patient at the right time. To achieve optimal outcomes treatment regimens now include complex decision-making processes surrounding the timing of chemotherapy and surgery. Recent research has demonstrated that some gastric cancer patients, such as those with tumours harbouring microsatellite instability, may be harmed by neoadjuvant chemotherapy. However, patients that achieve a good treatment response achieve superior clinical outcomes compared to adjuvant chemotherapy. Identifying specific subpopulations using tumour-based biomarkers is of critical importance to maximize outcomes.
Research objectives
We sought to characterize the expression of tumour immunohistochemistry (IHC)-based biomarkers CD4, CD8, Galectin-3 and E-cadherin in our Canadian population. Specifically, we evaluated these markers in comparison to their expression in normal gastric mucosa, as well as their relationship to neoadjuvant chemotherapy tumour response scores and expression in tumour biopsies before and after treatment. We successfully identified a biomarker, namely the CD4/CD8 T-cell ratio, with the potential to predict favourable treatment response. This pilot study serves as a foundation for future study to validate our preliminary findings.
Research methods
In this study, we evaluated IHC -based biomarkers in human gastric cancer specimens. Informed consent according to an approved ethics protocol was obtained for all patients. Samples were retrieved from endoscopic biopsy prior to treatment with neoadjuvant, adjuvant or palliative chemotherapy, as well as from pathology specimens following surgical resection. Using IHC, we quantified the expression of CD4, CD8, Galectin-3 and E-cadherin in gastric cancer tumours and adjacent normal mucosa. Quantification was performed on digitally scanned images using QuPath, which is an open-source and artificial intelligence-based digital pathology program. Statistical analysis was completed using R. Sample size calculations were performed using the MKpower package in R.
Research results
We demonstrate that an elevated CD4/CD8 ratio in gastric cancer tumours is significantly associated with complete or near complete response following FLOT chemotherapy. We identify that neoadjuvant chemotherapy is associated with increased infiltration of CD4 and CD8 T-cells in 15 paired samples assessed before and after exposure to chemotherapy. However, the dynamic increase in these lymphocyte populations does not associate with an increased CD4/CD8 ratio. To expand on the findings of this study, we performed a sample size calculation and identified that CD4, CD8, Galectin-3 and E-cadherin expression may be adequately evaluated with a future study population of 85 patients.
Research conclusions
For the first time, we identify that a high CD4/CD8 ratio within gastric cancer tumours is a promising biomarker that predicts favourable tumour response scores following neoadjuvant FLOT chemotherapy. To achieve this result, we use digital pathology technology and artificial intelligence-based quantification of biomarker staining.
Research perspectives
This study serves as a foundation for future research in validating the CD4/CD8 ratio as a reliable biomarker that is capable of predicting neoadjuvant treatment response. Our sample size calculations provide a framework for future study design.
FOOTNOTES
Author contributions: Skubleny D, McCall M, Ghosh S, Spratlin JL, Schiller D and Rayat G designed and coordinated the study; Skubleny D, Lin A, Garg S, McLean R performed experiments, generated and analyzed data; McLean R evaluated biopsy pathology and presence of cancer; Skubleny D, Ghosh S, Schiller D and Rayat G interpreted the data; Skubleny D wrote the manuscript with editorial assistance from Ghosh S, Spratlin JL, Schiller D and Rayat G.
Institutional review board statement:All human clinical participants consented according to the approved ethics protocol granted by the Health Research Ethics Board of Alberta (Study ID: HREBA.CC-17-0228_REN5).
Informed consent statement:Informed consent according to an approved ethics protocol from the Health Research Ethics Board of Alberta (HREBA.CC-17-0228) was obtained for all patients.
Conflict-of-interest statement:All authors have no conflicts of interest to disclose.
Data sharing statement:Raw data and code are available from the corresponding author at skubleny@ualberta.ca.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:Canada
ORCID number:Daniel Skubleny 0000-0001-8243-6139.
S-Editor:Liu GL
L-Editor:A
P-Editor:Yuan YY
 World Journal of Gastrointestinal Oncology2023年2期
World Journal of Gastrointestinal Oncology2023年2期
- World Journal of Gastrointestinal Oncology的其它文章
- Comment on “Crosstalk between gut microbiota and COVID-19 impacts pancreatic cancer progression”
- Prognostic value of claudin 18.2 expression in gastric adenocarcinoma
- Potent bromodomain and extraterminal domain inhibitor JAB-8263 suppresses MYC expression and exerts anti-tumor activity in colorectal cancer models
- microRNA-627-5p inhibits colorectal cancer cell proliferation,migration and invasion by targeting Wnt2
- Cancerous inhibitor of protein phosphatase 2A enhances chemoresistance of gastric cancer cells to oxaliplatin
- Evaluation of polygenic risk score for risk prediction of gastric cancer
