Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma
Zhao-Shan Niu, Wen-Hong Wang, Xian-Ning Dong, Li-Mei-Li Tian
Abstract Long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) are noncoding RNAs (ncRNAs) that occupy over 90% of the human genome, and their main function is to directly or indirectly regulate messenger RNA (mRNA) expression and participate in the tumorigenesis and progression of malignances. In particular, some lncRNAs can interact with miRNAs as competing endogenous RNAs (ceRNAs) to modulate mRNA expression. Accordingly, these RNA molecules are interrelated and coordinate to form a dynamic lncRNA-mediated ceRNA regulatory network. Mounting evidence has revealed that lncRNAs that act as ceRNAs are closely related to tumorigenesis. To date, numerous studies have established many different regulatory networks in hepatocellular carcinoma (HCC), and perturbations in these ceRNA interactions may result in the initiation and progression of HCC. Herein, we emphasize recent advances concerning the biological function of lncRNAs as ceRNAs in HCC, with the aim of elucidating the molecular mechanism underlying these HCC-related RNA molecules and providing novel insights into the diagnosis and treatment of HCC.
Key words: Hepatocellular carcinoma; Long noncoding RNA; MicroRNA; Competing endogenous RNA; Function; Mechanism
INTRODUCTION
Hepatocellular carcinoma (HCC) is a malignant tumor with high morbidity and mortality worldwide[1]. However, the pathogenesis of HCC remains elusive. Although great progress has been made in the diagnosis and treatment of HCC in recent years, the overall and long-term effects of treatment in patients with advanced HCC are poor. Therefore, in-depth studies are needed to explore the mechanisms underlying HCC occurrence and development, which will contribute to the development of effective diagnostic biomarkers and therapeutic targets for HCC.
Protein-coding genes account for less than 2% of the human genome, while most of the genome is composed of genes that are transcribed into noncoding RNAs (ncRNAs)[2]. NcRNAs are divided into long noncoding RNAs (lncRNAs), small ncRNAs, and intermediate-sized ncRNAs by length[3]. LncRNAs have been identified as key regulators of transcription and translation and are involved in a variety of biological processes by regulating gene expression[4]. MicroRNAs (miRNAs) are small ncRNAs that interact with the 3’-untranslated region (3’-UTR) of target mRNAs to facilitate their degradation or inhibit their translation. MiRNAs play a critical role in tumorigenesis and tumor cell proliferation, migration, and invasion[5]. LncRNAs and miRNAs are regulatory ncRNAs, and dysregulation of lncRNAs or miRNAs is involved in tumor initiation and progression eitherviathe activation or inhibition of target genes[6].
Existing evidence indicates that there are interactions among RNA molecules, such as lncRNAs and miRNAs[7], miRNAs and mRNAs[8], and lncRNAs and mRNAs[9]; these RNA molecules are interrelated and collaborate to form a dynamic regulatory network of lncRNAs acting as competitive endogenous RNAs (ceRNAs)[10]. The ceRNA mechanism is one of the important ways by which an lncRNA exerts its posttranscriptional gene regulation in the cytoplasm, and perturbations in these ceRNA interactions contribute to tumor initiation and progression. Currently, the identified ceRNAs include protein-coding RNAs (mRNAs) and ncRNAs, such as lncRNAs, pseudogene transcripts, viral RNAs, and circular RNAs (circRNAs). LncRNAs are the main component of the ceRNA network, as they regulate mRNA expression by acting as miRNA sponges. To date, numerous different regulatory networks of lncRNAs acting as ceRNAs in HCC have been established. Accumulating evidence has revealed that lncRNAs acting as ceRNAs play pivotal roles in HCC initiation and progression[11,12]. Herein, we emphasize recent advances concerning the biological function of lncRNAs acting as ceRNAs in HCC, with the aim of elucidating the molecular mechanism underlying these HCC-related RNA molecules and providing novel insights into the diagnosis and treatment of HCC.
MECHANISM OF ACTION OF LNCRNAS INVOLVED IN THE CERNA REGULATORY NETWORK
Theoretically, any RNA molecule with a miRNA binding site can bind to a miRNA to form an intricate ceRNA regulatory network. RNA transcripts in the ceRNA network are in a state of equilibrium under physiological conditions; once perturbed, this will lead to the occurrence of disease[13,14]. In the ceRNA regulatory network, miRNA acts as a bridge between ncRNA and mRNA and negatively regulates the expression of its target mRNA[15]. There is growing evidence that each miRNA can regulate many transcripts. In turn, RNA transcripts with different miRNA response elements (MREs) may also be targets of multiple miRNAs[16]. The multiplicity of targets allows RNA and RNA to interact with each other by competitively binding to a common MRE, and the same MRE is the structural basis for the binding of different RNAs[16].
In addition to directly regulating mRNAs, lncRNAs can also indirectly affect the expression of target genes by sponging miRNAs[10]. Structurally, most lncRNAs are similar to mRNAs, which makes their patterns of gene regulation more diverse and extensive and unaffected by translation[17]. This may be the reason why many lncRNAs can act as ceRNAs to sponge miRNAs to inhibit miRNAs from degrading their target mRNAs. In general, the more miRNA binding sites there are on an lncRNA, the stronger the competition[18]. When lncRNAs are expressed at low levels, they can bind only a few miRNAs, and the remaining miRNAs interact with mRNAs to promote their degradation. In contrast, when lncRNAs are expressed at high levels, they can combine with more miRNAs, thus relieving the inhibitory effect of miRNAs on their target mRNAs. The new regulatory pattern of lncRNA-miRNA-mRNA is an extension of the traditional miRNA-mRNA regulatory model[10].
ROLE OF LNCRNAS AS CERNAS IN HCC
To date, mounting evidence indicates that oncogenic or tumor suppressive lncRNAs can regulate their target genes by acting as ceRNAs to sponge miRNAs[19,20], thereby affecting glucose metabolism, immune escape, autophagy, angiogenesis, liver cancer stem cells (LCSCs), proliferation, apoptosis, epithelial-mesenchymal transition (EMT), migration, invasion, metastasis, chemoresistance, and radioresistance in HCC (Figure 1). Specifically, the majority of the identified lncRNAs exhibit oncogenic properties that function as ceRNAs for tumor suppressive miRNAs, thereby activating the expression of oncogenic mRNAs to promote HCC occurrence and progression (Figure 2A). In addition, some lncRNAs exhibit tumor suppressive properties, acting as ceRNAs for oncogenic miRNAs (oncomiRs), thus upregulating the expression of tumor suppressive targets to inhibit HCC occurrence and progression (Figure 2B). Here, we elucidate the functions of some lncRNA-mediated ceRNA regulatory networks in HCC (Table 1). Note that there are many abbreviations in this paper, so they are listed and expanded in Table 1 below.
Glucose metabolism
The “reprogramming” of glucose metabolism is regarded as a prominent characteristic of cancer cells. A large amount of lactic acid produced by glycolysis forms an inflammatory microenvironment around the tumor, which contributes to tumor cell proliferation, EMT, invasion, metastasis, immune escape, and resistance to chemotherapy and radiotherapy. Existing evidence indicates that aberrant glucose metabolism plays a pivotal role in the invasion and metastasis of HCC[21,22]. The mechanism of abnormal activation of glycolysis in cancer cells is complex, and many studies have confirmed that lncRNAs play a significant role in modulating glycolysis by sponging miRNAs in HCC, among which oncogenic lncRNAs that act as ceRNAs can promote glycolysis. For instance, lactate dehydrogenase isoform A (LDHA), a glycolytic enzyme, can mediate aerobic glycolysis in cancer cells[23]. The lncRNARAET1K, as a miR-100-5p sponge, can enhance LDHA expression and facilitate hypoxia-induced glycolysis, thereby promoting HCC progression[24]. In addition, hemikinase 2 (HK2) is another glycolytic enzyme related to glycolysis in cancer cells[25], and the lncRNATUG1induces glycolysis and promotes HCC metastasis by acting as a ceRNA to enhance HK2 expression by sponging miR-455-3p[26]. Additionally, hypoxia-inducible factor (HIF) 1 has been confirmed to promote aerobic glycolysis in cancer[27], and the lncRNAHOTAIRpromotes glycolysis by acting as a ceRNA for miR-130a-3p to increase HIF1 expression in HCC cells[28]. By contrast, tumor suppressive lncRNAs that act as ceRNAs can inhibit glycolysis in HCC. For example, endoplasmic reticulum protein 29 (ERp29), an endoplasmic reticulum protein, and the lncRNAMEG3are downregulated in high-glucose (HG) HCC cells, while miR-483-3p is upregulated in HG HCC cells. Mechanistically, the overexpression ofMEG3inhibits glycolysis by sponging miR-483-3p to increase ERp29 expression in HCC[29]. Currently, antitumor drugs that target glucose metabolism are being researched and developed;the above findings suggest that the lncRNA-mediated ceRNA network could provide new ideas for inhibiting glycolysis in HCC.
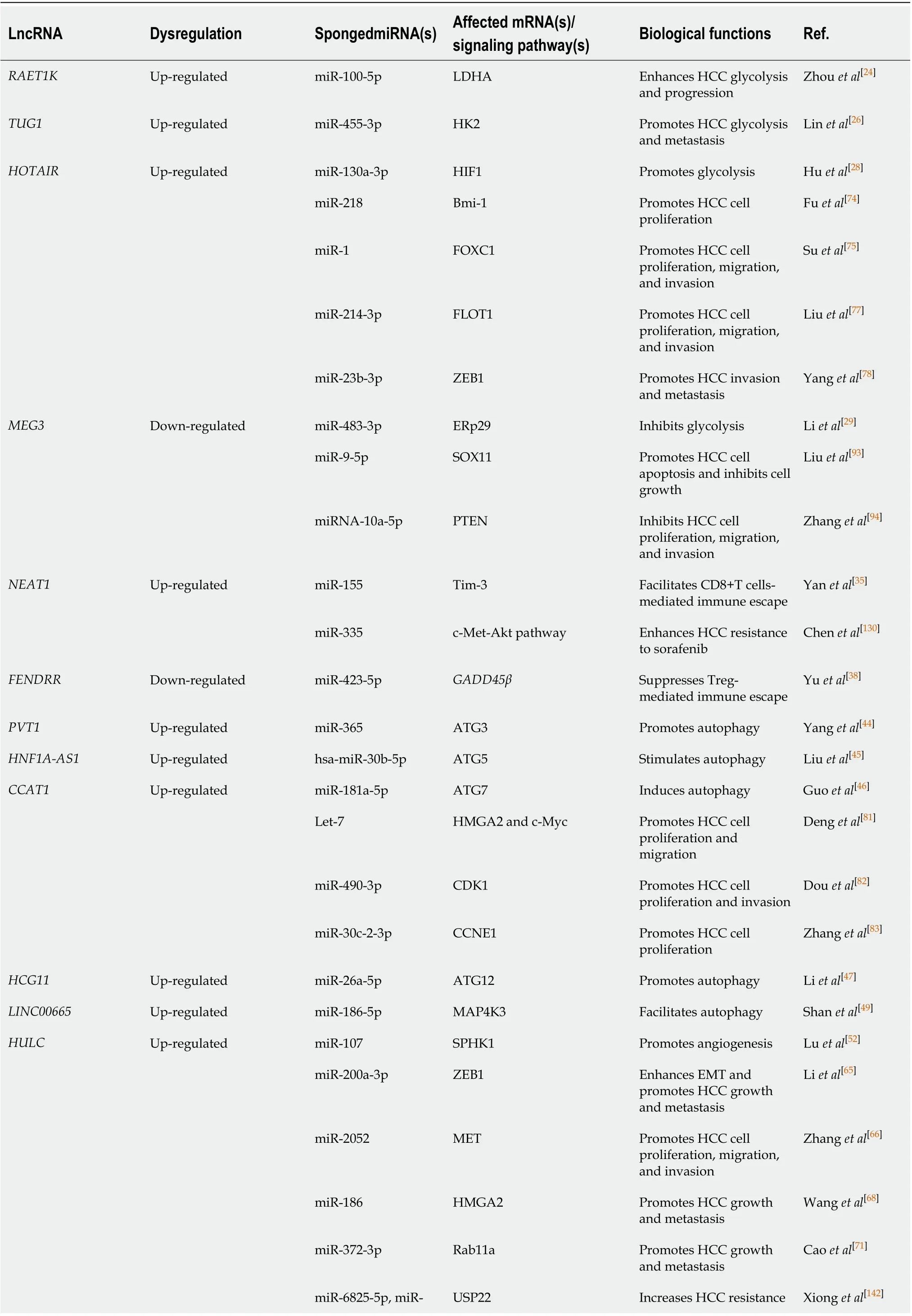
Table 1 Long noncoding RNA-mediated competitive endogenous RNA network in hepatocellular carcinoma
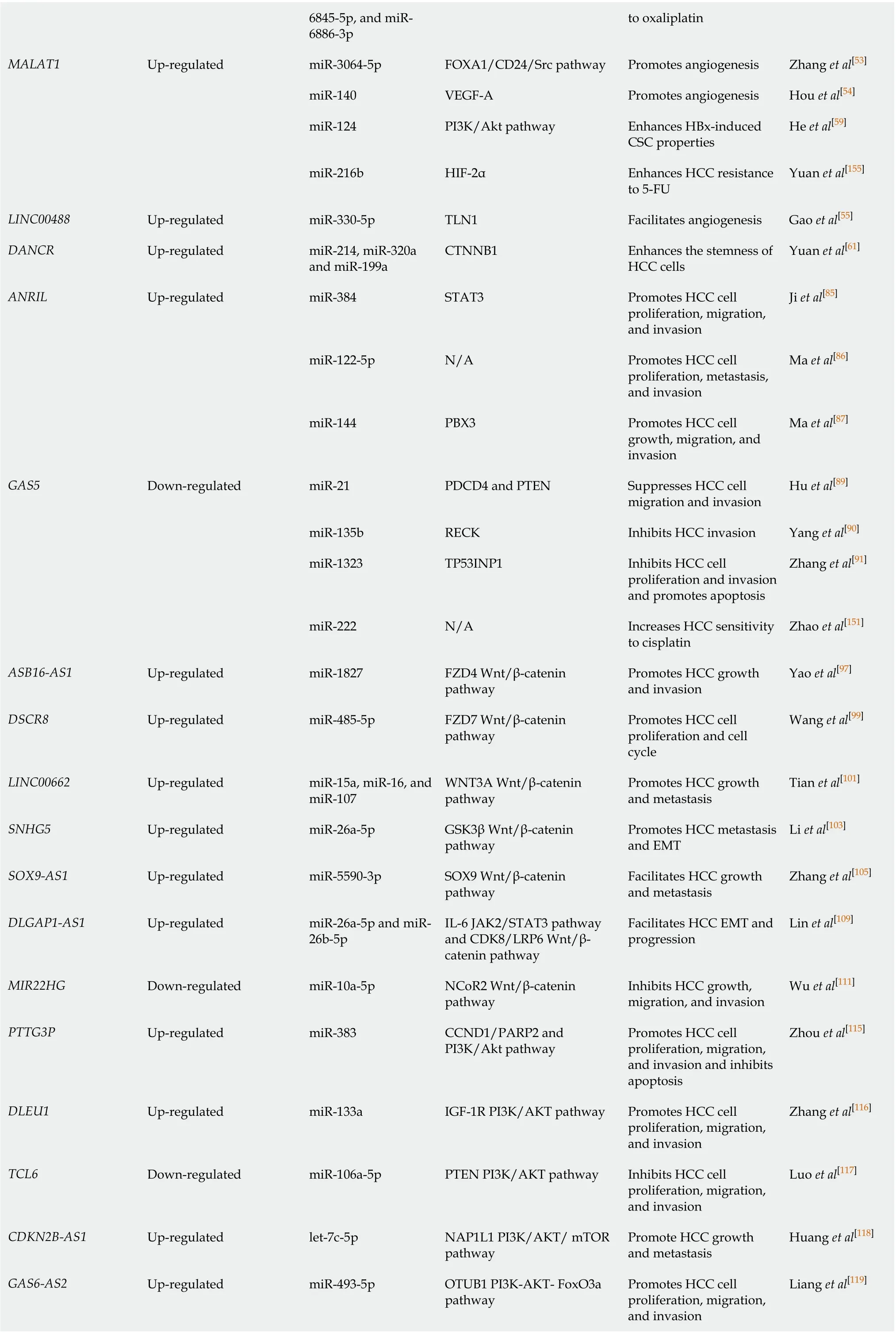
6845-5p, and miR-6886-3p to oxaliplatin MALAT1 Up-regulated miR-3064-5p FOXA1/CD24/Src pathway Promotes angiogenesis Zhang et al[53]miR-140 VEGF-A Promotes angiogenesis Hou et al[54]miR-124 PI3K/Akt pathway Enhances HBx-induced CSC properties He et al[59]miR-216b HIF-2α Enhances HCC resistance to 5-FU Yuan et al[155]LINC00488 Up-regulated miR-330-5p TLN1 Facilitates angiogenesis Gao et al[55]DANCR Up-regulated miR-214, miR-320a and miR-199a CTNNB1 Enhances the stemness of HCC cells Yuan et al[61]ANRIL Up-regulated miR-384 STAT3 Promotes HCC cell proliferation, migration, and invasion Ji et al[85]miR-122-5p N/A Promotes HCC cell proliferation, metastasis, and invasion Ma et al[86]miR-144 PBX3 Promotes HCC cell growth, migration, and invasion Ma et al[87]GAS5 Down-regulated miR-21 PDCD4 and PTEN Suppresses HCC cell migration and invasion Hu et al[89]miR-135b RECK Inhibits HCC invasion Yang et al[90]miR-1323 TP53INP1 Inhibits HCC cell proliferation and invasion and promotes apoptosis Zhang et al[91]miR-222 N/A Increases HCC sensitivity to cisplatin Zhao et al[151]ASB16-AS1 Up-regulated miR-1827 FZD4 Wnt/β-catenin pathway Promotes HCC growth and invasion Yao et al[97]DSCR8 Up-regulated miR-485-5p FZD7 Wnt/β-catenin pathway Promotes HCC cell proliferation and cell cycle Wang et al[99]LINC00662 Up-regulated miR-15a, miR-16, and miR-107 WNT3A Wnt/β-catenin pathway Promotes HCC growth and metastasis Tian et al[101]SNHG5 Up-regulated miR-26a-5p GSK3β Wnt/β-catenin pathway Promotes HCC metastasis and EMT Li et al[103]SOX9-AS1 Up-regulated miR-5590-3p SOX9 Wnt/β-catenin pathway Facilitates HCC growth and metastasis Zhang et al[105]DLGAP1-AS1 Up-regulated miR-26a-5p and miR-26b-5p IL-6 JAK2/STAT3 pathway and CDK8/LRP6 Wnt/βcatenin pathway Facilitates HCC EMT and progression Lin et al[109]MIR22HG Down-regulated miR-10a-5p NCoR2 Wnt/β-catenin pathway Inhibits HCC growth, migration, and invasion Wu et al[111]PTTG3P Up-regulated miR-383 CCND1/PARP2 and PI3K/Akt pathway Promotes HCC cell proliferation, migration, and invasion and inhibits apoptosis Zhou et al[115]DLEU1 Up-regulated miR-133a IGF-1R PI3K/AKT pathway Promotes HCC cell proliferation, migration, and invasion Zhang et al[116]TCL6 Down-regulated miR-106a-5p PTEN PI3K/AKT pathway Inhibits HCC cell proliferation, migration, and invasion Luo et al[117]CDKN2B-AS1 Up-regulated let-7c-5p NAP1L1 PI3K/AKT/ mTOR pathway Promote HCC growth and metastasis Huang et al[118]GAS6-AS2 Up-regulated miR-493-5p OTUB1 PI3K-AKT- FoxO3a pathway Promotes HCC cell proliferation, migration, and invasion Liang et al[119]
HCC: Hepatocellular carcinoma; LncRNAs: Long noncoding RNAs; ceRNA: Competitive endogenous RNA; RAET1K: Retinoic acid early transcript 1K; LDHA: Lactate dehydrogenase isoform A; TUG1: Taurine up-regulation gene 1; HK2: Hemikinase 2; HOTAIR: Homeobox transcript antisense RNA; HIF1: Hypoxia-inducible factor 1; Bmi-1:B lymphoma moloney murine leukemia virus insertion region 1; FOXC1: Forkhead box C1; FLOT1: Flotillin 1; ZEB1: Zinc finger E-box binding homeobox 1; MEG3: Maternally expressed gene 3; ERp29: ER protein 29; SOX11: SRY-related HMG-box transcription factor 11; PTEN: Phosphatase and tensin homolog; NEAT1: Nuclear enriched abundant transcript 1; Tim-3: T cell immunoglobulin mucin-3; FENDRR: Fetal-lethal noncoding developmental regulatory RNA; GADD45β: Growth arrest and DNA damage-inducible beta; PVT1: Plasmacytoma variant translocation 1; ATG3: Autophagy related genes 3; HNF1A-AS1: HNF1A antisense RNA 1; ATG5: Autophagy related genes 5; CCAT1: Colon cancer associated transcript 1; ATG7: Autophagy related genes 7; HMGA2: High mobility group AT-hook 2; CDK1: Cyclin-dependent kinase 1; CCNE1: Cyclin E1; HCG11: HLA complex group 11; ATG12: Autophagy related genes 12; LINC00665: Long intergenic non-protein coding RNA 665; MAP4K3: Mitogen-activated protein kinase kinase kinase kinase 3; HULC: Highly upregulated in liver cancer; SPHK1: Sphingosine kinase 1; MET: Hepatocyte growth factor receptor; Rab11a: Member RAS oncogene family; USP22: Ubiquitin-specific peptidase 22; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; FOXA1: Forkhead box A1; VEGF-A: Vascular endothelial growth factor A; PI3K/Akt pathway: Phosphoinositide-3-kinase/protein kinase B; CSC: Liver cancer stem cell; HIF-2α: Hypoxia-inducible factor 2α; TLN1: Talin 1; DANCR: Differentiation antagonizing non-protein coding RNA; CTNNB1: Catenin beta-1; ANRIL: Antisense noncoding RNA in the INK4 locus; STAT3: Signal transducer and activator of transcription 3; N/A: Not available.; PBX3: Pre-B-cell leukemia homeobox 3; GAS5: Growth arrest-specific 5; PDCD4: Programmed cell death 4; RECK: Cysteine-rich protein with Kazal motifs; TP53INP1:Tumor protein p53-induced nuclear protein 1;ASB16-AS1:ASB16 antisense RNA 1; FZD4: Frizzled 4; DSCR8: Down syndrome critical region 8; FZD7: Frizzled-7; WNT3A:Wingless-type MMTV integration site family 3A; SNHG5: Small nucleolar RNA host gene 5; GSK3β: Glycogen synthase kinase 3β; EMT: Epithelial-mesenchymal transition; SOX9-AS1: SOX9 antisense RNA 1; SOX9: Sex determining region Y-box 9; DLGAP1-AS1: Long noncoding RNA DLGAP1 antisense RNA 1; IL-6: Interleukin- 6; JAK2: Janus kinase 2; CDK8: Cyclin-dependent kinase 8; LRP6: Low-density lipoprotein receptorrelated protein 6; MIR22HG: MIR22 host gene; NCoR2: Nuclear receptor corepressor 2; PTTG3P: Pituitary tumor-transforming 3; CCND1: Cyclin D1; PARP2: Poly ADP-ribose polymerase 2; DLEU1: Deleted in lymphocytic leukaemia 1; IGF-1R : Insulin-like growth factor 1 receptor; TCL6: T cell leukemia/lymphoma 6; CDKN2B-AS1: CDKN2B antisense RNA 1; NAP1L1: Nucleosome assembly protein 1 like 1; mTOR: Mammalian rapamycin; GAS6-AS2: Growth arrest specific 6 antisense RNA 2; OTUB1: OTU domain-containing ubiquitin aldehyde-binding protein 1; FOXO3a: Forkhead Box O3a; SNHG12: Small nucleolar RNA host gene 12; MLK3: Mixed-lineage kinase 3; NF-kB: Nuclear factor kappa-B; CASC2: Cancer susceptibility candidate 2; CYLD: Cylindromatosis; LINC-ROR: Intergenic non-protein coding RNA, regulator of reprogramming; FoxM1: Forkhead box M1; RAD18: A RING-type ubiquitin ligase E3; PIK3R3: Phosphoinositide-3-kinase regulatory subunit 3; FOXD2-AS1: FOXD2 adjacent opposite strand RNA 1; TMEM9: Transmembrane protein 9; SNHG3: Small nucleolar RNA host gene 3; SNHG16: Small nucleolar RNA host gene 16; NR2F1-AS1: NR2F1 antisense RNA 1; ABCC1: Multidrug resistance-associated protein 1; KCNQ1OT1: KCNQ1 overlapping transcript 1; NRAL: Nrf2 regulation-associated lncRNA; Nrf2: Nuclear factor erythroid-2-related factor 2; MAGEA3: Melanoma-associated antigen A3; CRNDE: Colorectal neoplasia differentially expressed; KRAL: Keap1 regulation-associated lncRNA; Keap1: Kelch-like ECH-associated protein 1; NEAT1_2: Nuclear enriched abundant transcript 1_2; WEE1: WEE1 G2 checkpoint kinase; FOXP1: Forkhead box protein P1.
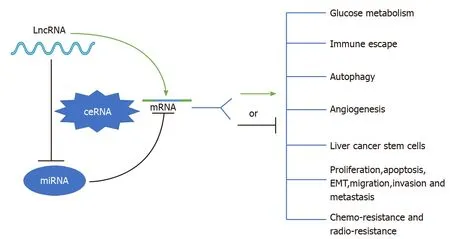
Figure 1 Schematic diagram of the role of long noncoding RNA-mediated competitive endogenous RNA regulatory network in hepatocellular carcinoma. See the text for details. LncRNA: Long noncoding RNA; ceRNA: Competing endogenous RNA; miRNA: MicroRNA; mRNA: Messenger RNA; EMT: Epithelial-mesenchymal transition.
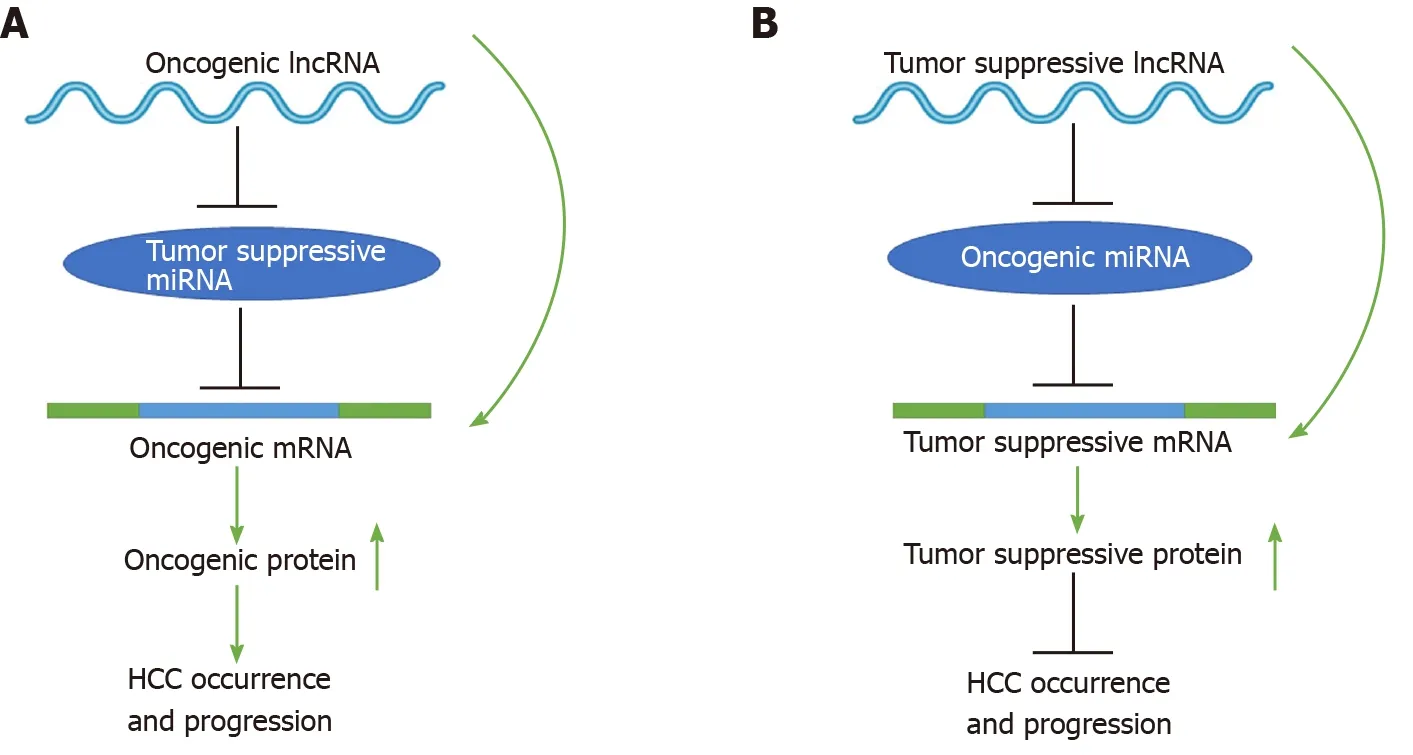
Figure 2 Schematic diagrams of long noncoding RNA-mediated competitive endogenous RNA regulatory network that mediates the occurrence and progression of hepatocellular carcinoma. See the text for details. LncRNA: Long noncoding RNA; miRNA: MicroRNAs; mRNA: Messenger RNA.
Immune escape
Tumor immune escape refers to the phenomenon that tumor cells can survive and proliferate by escaping immune system-mediated recognition and attack by changing themselves or their tumor microenvironment[30]. Currently, the effectiveness of immunotherapy is limited by tumor immune escape. Thus, an in-depth exploration of the mechanisms of tumor immune escape may provide novel insights into tumor immunotherapy. Current studies have shown that lncRNAs can modulate immune escape in HCC by acting as ceRNAs of miRNAs, among which oncogenic lncRNAs that function as ceRNAs can promote the immune escape of HCC cells. For example,NEAT1, a newly discovered oncogenic lncRNA, is specifically localized in nuclear paraspeckles and participates in paraspeckle formation and the transcriptional regulation of many genes[31,32]. T cell immunoglobulin mucin-3 (Tim-3), an immune checkpoint molecule, can suppress the immune response[33], and the increased expression of Tim-3 within the tumor can inactivate killer T cells, thus preventing the death of tumor cells[34]. Mechanistically,NEAT1facilitates the CD8+ T cell-mediated immune escape of HCC cells by acting as a ceRNA for miR-155 to enhance Tim-3 expression[35]. Conversely, tumor suppressive lncRNAs that act as ceRNAs can inhibit the immune escape of HCC cells. For instance,GADD45β, a tumor suppressor, is associated with antitumor immune responses[36], and CD4+T cells lackingGADD45βare less responsive to the stimulation of T cell receptors or inflammatory cytokines[37].FENDRR, a tumor suppressor lncRNA, upregulatesGADD45βby sponging miR-423-5p, thereby suppressing the immune escape of HCC cells[38]. These findings suggest that the lncRNA-mediated ceRNA network is involved in mediating immune evasion in HCC and thus may be a promising therapeutic target for HCC immunotherapy.
Autophagy
Autophagy is closely associated with the development of malignant tumors and can promote tumor survival and proliferation by regulating interactions between the tumor and tumor microenvironment[39]. In HCC, autophagy plays a vital role in tumor immunity, oxidative stress, and the maintenance of hepatic homeostasis and thus participates in HCC initiation and progression and resistance to chemotherapy drugs[40]. Identification of the mechanisms by which autophagy is activated in HCC will help clarify HCC pathogenesis and reveal novel treatments for HCC patients. Numerous investigations have indicated that oncogenic lncRNAs that function as ceRNAs are required for promoting autophagy in HCC. For example, autophagyrelated genes 3, 5, 7, and 12 (ATG3,ATG5,ATG7, andATG12, respectively) are major regulators of the induction of autophagosome formation[41-43]. The lncRNAPVT1promotes autophagy in HCC by enhancingATG3expression by sponging miR-365[44]. The lncRNAHNF1A-AS1upregulates the expression ofATG5in HCC by acting as a sponge of hsa-miR-30b-5p, thus stimulating autophagy[45]. The lncRNACCAT1serves as a ceRNA for miR-181a-5p to induce autophagy in HCC by enhancingATG7expression[46]. The lncRNAHCG11promotes autophagy in HCC by enhancingATG12expression by sponging miR-26a-5p[47]. In addition, mitogen-activated protein kinase kinase kinase kinase 3 (MAP4K3), an upstream kinase of the MAPK pathway, is a key node in the regulation of autophagy[48]. The lncRNALINC00665facilitates autophagy by sponging miR-186-5p to enhance MAP4K3 expression[49]. Collectively, these results suggest that the lncRNA-mediated ceRNA network could provide novel treatments for HCC patients.
Angiogenesis
Angiogenesis is responsible for HCC growth, proliferation, invasion, and metastasis[50]. The mechanisms underlying HCC angiogenesis are complex, and exploration of the factors involved in regulating HCC angiogenesis is of great significance for improving antiangiogenic treatments for HCC. Emerging evidence indicates that oncogenic lncRNAs that act as ceRNAs are tightly linked to HCC angiogenesis. For instance, sphingosine kinase 1 (SPHK1), a key metabolic enzyme, is correlated with tumor angiogenesis[51]. A study found that the lncRNAHULCpromotes angiogenesis by upregulating SPHK1 in HCC;HULCacts as a ceRNA to increase the expression of transcription factor E2F1 by competitively binding to miR-107 and subsequently results in the activation of theSPHK1promoter, thus promoting HCC angiogenesisin vivo[52]. In addition, the lncRNAMALAT1can promote HCC angiogenesis by sponging miR-3064-5p to activate the forkhead box A1 (FOXA1)/CD24/Src pathway[53]or by functioning as a miR-140 sponge to enhance vascular endothelial growth factor A expression[54]. Similarly,LINC00488, another lncRNA, upregulates the expression of talin 1 to facilitate HCC angiogenesis by sponging miR-330-5p[55]. These findings suggest that the lncRNA-mediated ceRNA network may be a promising target for antiangiogenic therapies for HCC.
Liver cancer stem cells
LCSCs exhibit high proliferation, self-renewal, high tumorigenicity, chemoresistance, and radioresistance[56-58], and their abundance is positively associated with the degree
of HCC malignancy. Elucidation of the regulatory mechanisms of LCSCs will contribute to our understanding of the pathogenesis of HCC and the identification of novel therapeutic strategies. Existing evidence has shown that oncogenic lncRNAs help sustain cancer stem cell (CSC) traits by acting as ceRNAs for miRNAs to initiate HCC development. For instance, the lncRNAMALAT1activates the phosphoinositide-3-kinase/protein kinase B (PI3K/Akt) pathway by acting as a miR-124 sponge to enhance HBx-induced CSC properties[59]. In addition, catenin beta-1 (CTNNB1)/βcatenin sustains the stemness properties of LCSCs[60]. In another recent study, it was found that the lncRNADANCRwas highly expressed in HCC tissues and stem-like HCC cells;DANCRcan act as a ceRNA to enhance CTNNB1 expression by sponging miR-214, miR-320a, and miR-199a, thereby enhancing the stemness of HCC cells[61]. Thus, the lncRNA-mediated ceRNA network may serve as a potential therapeutic target for LCSCs.
Proliferation, migration, EMT, invasion, and metastasis
The lncRNA-mediated ceRNA regulatory network functions by regulating miRNA target genes:LncRNAs acting as ceRNAs can participate in cell proliferation, apoptosis, migration, EMT, invasion, and metastasis by modulating mRNAs in HCC. Currently, there are many lncRNA-miRNA-miRNA target gene (mRNA) networks reported in HCC. In this review, we provide only a few examples of lncRNA-miRNAmRNA networks, including those involving oncogenic lncRNAs such asHULC, HOTAIR, CCAT, andANRILand tumor suppressive lncRNAs such asGAS5andMEG3.
LncRNA HULC-miRNA-mRNA:HULChas been identified as a specifically highly expressed lncRNA in HCC[62]. In a recent study, highHULCexpression in HCC was significantly connected to increased lymph node metastasis and advanced TNM stage[63]. This finding indicates thatHULCcan facilitate the proliferation, migration, invasion, and metastasis of HCC cells, leading to the malignant development of HCC. Thus far, it has been reported thatHULCmay exert its oncogenic function in HCC through diverse molecular mechanisms, of which theHULC-mediated ceRNA network is important. For instance, zinc finger E-box binding homeobox 1 (ZEB1), a key regulator of EMT, contributes to HCC cell invasion and metastasis[64]. As a ceRNA of miR-200a-3p,HULCincreases the expression of ZEB1, thereby enhancing EMT and promoting HCC growth and metastasis[65]. In addition,HULCcan enhance the expression of hepatocyte growth factor receptor (MET) by sponging miR-2052, thus promoting HCC cell proliferation, migration, and invasion[66]. High mobility group AT-hook 2 (HMGA2), an oncogene, has been shown to be closely associated with cancer progression and metastasis[67].HULCpromotes HCC growth and metastasis by enhancing HMGA2 expression by acting as a ceRNA of miR-186[68]. Rablla, a central regulatory protein, promotes exosome secretion, and exosomes significantly promote HCC progression[69,70]. A mechanistic investigation revealed thatHULCincreases RAS oncogene family member expression to induce exosome secretion by sponging miR-372-3p, contributing to HCC growth and metastasis[71].
LncRNA HOTAIR-miRNA-mRNA:The lncRNAHOTAIRhas been reported to exert an oncogenic role in a variety of malignances[72,73]. Emerging evidence suggests thatHOTAIRfunctions as a ceRNA and facilitates HCC cell proliferation, migration, invasion, and metastasis. For instance, a study confirmed thatHOTAIRpromotes HCC cell proliferation and tumorigenicity by competitively binding to miR-218 to activate B lymphoma Moloney murine leukemia virus insertion region 1 expression and inactivate P16Ink4aand P14ARF[74]. Another study also demonstrated that Forkhead box C1-activatedHOTAIRpromotes HCC cell proliferation, migration, and invasion by acting as a sponge of miR-1[75]. In addition, flotillin 1 (FLOT1), a marker of lipid rafts, is highly expressed in HCC and contributes to aggressive tumor characteristics[76].HOTAIRenhances FLOT1 expression by sponging miR-214-3p, thereby promoting HCC cell proliferation, migration, and invasion[77]. Additionally,HOTAIRpromotes HCC cell invasion and metastasis by sponging miR-23b-3p to upregulate ZEB1 expression[78].
LncRNA CCAT-miRNA-mRNA:The lncRNACCAT1is located on chromosome 8q24.21 and plays vital roles in promoting HCC cell proliferation and metastasis[79].CCAT1has been shown to upregulate the expression of its downstream genec-Myc, thereby promoting tumorigenesis[80]. Subsequently, many studies have explored the potential mechanism by whichCCAT1upregulatesc-Mycto promote tumorigenesis. In HCC,CCAT1functions as a ceRNA of miRNA let-7, thus counteracting the inhibitory effect of Let-7 on its target genes,HMGA2andc-Myc, which upregulates the expression ofHMGA2andc-Mycand ultimately facilitates HCC proliferation and migration[81]. In addition,CCAT1upregulates cyclin-dependent kinase 1 expression by acting as a miR-490-3p sponge, thereby promoting HCC cell proliferation and invasion[82]. Furthermore,CCAT1acts as a sponge of miR-30c-2-3p to upregulate the expression of cyclin E1, leading to HCC cell proliferation[83].
LncRNA ANRIL-miRNA-mRNA:The lncRNAANRILis located on chromosome 9p21 and plays an oncogenic role in tumorigenesis[84].ANRILfunctions as a ceRNA to sponge miRNAs, thereby regulating gene expression in HCC. The high expression ofANRILin HCC is related to HCC cell proliferation, migration, and invasion; mechanistically,ANRILexerts its biological action in HCC by sponging miR-384 to upregulate signal transducer and activator of transcription 3 (STAT3) expression[85].ANRILcan also promote HCC cell proliferation, metastasis, and invasion by acting as a ceRNA of miR-122-5p[86]. In addition, ANRIL can upregulate the expression of pre-Bcell leukemia homeobox 3 by sponging miR-144 to facilitate HCC cell growth, migration, and invasion[87].
LncRNA GAS5-miRNA-mRNA:The lncRNAGAS5is downregulated in diverse malignancies, including HCC[88]. Increasing evidence indicates that theGAS5-mediated ceRNA network may be one of the important mechanisms by whichGAS5exerts its biological functions in HCC. For example,GAS5restrains HCC cell migration and invasion by sponging miR-21 to upregulate its targets, programmed cell death 4 and phosphatase and tensin homolog (PTEN)[89]. In addition,GAS5suppresses HCC invasion by sponging miR-135b to enhance cysteine-rich protein with Kazal motifs expression[90].GAS5also functions as a miR-1323 sponge to upregulate tumor protein p53-induced nuclear protein 1 expression, thus inhibiting HCC cell proliferation and invasion and promoting apoptosis[91].
LncRNA MEG3-miRNA-mRNA:MEG3is an imprinted gene and a tumor suppressive lncRNA.MEG3is inversely related to tumorigenesis and plays an inhibitory role in many malignancies[92]. Acting as a ceRNA against miRNA is an important mechanism of action ofMEG3in HCC. For example, the overexpression ofMEG3can promote cell apoptosis and inhibit HCC growth by upregulating SRYrelated HMG-box transcription factor 11 expression by acting as an miR-9-5p sponge[93]. In addition,MEG3enhances the expression of PTEN to restrain HCC cell proliferation, migration, and invasion by sponging miRNA-10a-5p[94].
The lncRNA-mediated ceRNA regulatory network functions by modulating signaling pathways:LncRNAs acting as ceRNAs can also participate in HCC cell proliferation, migration, EMT, invasion, and metastasis by modulating various signaling pathways in HCC, including the Wnt/β-catenin pathway, PI3K/AKT pathway, and nuclear factor kappa-B (NF-kB) pathway.
Wnt/β-catenin pathway:Abnormal activation of the Wnt/β-catenin pathway, a key event implicated in HCC carcinogenesis, is believed to be a key target for the clinical diagnosis and treatment of HCC[95]. Thus, elucidation of the regulatory mechanisms of the Wnt/β-catenin pathway will provide new insights into a new anticancer therapy for HCC. Extensive evidence to date has indicated that lncRNAs can mediate the Wnt/β-catenin pathway by acting as ceRNAs of miRNAs, thereby modulating HCC cell proliferation and invasion; oncogenic lncRNAs that act as ceRNAs can perform their biological actions by activating the Wnt/β-catenin pathway in HCC. For example, frizzled (FZD) 4, a Wnt receptor, can activate the Wnt/β-catenin pathway in HCC[96], and the lncRNAASB16-AS1enhances FZD4 expression to activate the Wnt/βcatenin pathway by acting as a miR-1827 sponge and subsequently facilitates HCC growth and invasion[97]. Likewise, another Wnt receptor, FZD7, can also activate the Wnt/β-catenin pathway in HCC[98]. The lncRNADSCR8activates the Wnt/β-catenin pathway by enhancing FZD7 expression by acting as a miR-485-5-p sponge to facilitate HCC cell proliferation and the cell cycle[99]. Wingless-type MMTV integration site family 3A (WNT3A) is one of the crucial components of the Wnt/β-catenin pathway related to HCC progression[100], and the lncRNALINC00662activates the Wnt/βcatenin pathway by enhancing WNT3A expressionviathe competitive sponging of miR-15a, miR-16, and miR-107, thereby promoting HCC growth and metastasis[101]. Glycogen synthase kinase 3β (GSK3β) is a pivotal regulator of β-catenin signaling[102], and the lncRNASNHG5acts as a miR-26a-5p sponge to enhance GSK3β expression, thereby activating the Wnt/β-catenin pathway to facilitate HCC metastasis and EMT[103]. In addition, sex determining region Y-box (SOX) 9 can activate the Wnt/βcatenin pathway in HCC[104], and the lncRNA SOX9-AS1facilitates HCC growth and metastasis by increasing SOX9 expression to activate the Wnt/β-catenin pathway by acting as a miR-5590-3p sponge[105]. In HCC, interleukin (IL)-6 is associated with the activation of Janus kinase 2 (JAK2)/STAT3 signaling[106]; cyclin-dependent kinase (CDK) 8 and low-density lipoprotein receptor-related protein 6 (LRP6) are associated with the activation of Wnt/β-catenin signaling[107,108], and the lncRNADLGAP1-AS1increases the expression of IL-6 and CDK8/LRP6 by functioning as a sponge of miR-26a-5p and miR-26b-5p, thereby activating JAK2/STAT3 and Wnt/β-catenin signaling to facilitate EMT and the progression of HCC, respectively[109]. Instead, tumor suppressive lncRNAs that act as ceRNAs function by inactivating Wnt/β-catenin signaling in HCC. For example, in HCC, nuclear receptor corepressor 2 is associated with inhibition of the activation of Wnt/β-catenin signaling[110].MIR22HG, a tumor suppressive lncRNA, increases NCOR2 expression by sponging miR-10a-5p, thereby inactivating Wnt/β-catenin signaling to inhibit HCC cell growth, migration, and invasion[111]. The abovementioned findings suggest that different lncRNA-mediated ceRNA networks can exert their biological functions in HCC by mediating the Wnt/βcatenin pathway; these networks may become effective therapeutic targets for treating HCC patients.
PI3K/AKT pathway:PI3K/AKT, a highly activated pathway in HCC, is implicated in HCC carcinogenesis and chemoresistance[112-114]. At present, emerging evidence indicates that multiple lncRNA-mediated ceRNA networks can exert their biological functions by modulating the PI3K/AKT pathway, among which oncogenic lncRNAs that act as ceRNAs exert their biological function by activating the PI3K/AKT pathway in HCC. For instance, the lncRNAPTTG3Pfacilitates the proliferation, migration, and invasion and inhibits the apoptosis of HCC cells by increasing the expression of cyclin D1/poly ADP-ribose polymerase 2 and activating the PI3K/Akt pathway by acting as a ceRNA of miR-383[115]. Similarly, the lncRNADLEU1activates the PI3K/Akt pathway by increasing insulin-like growth factor 1 receptor-1R expression by sponging miR-133a, thereby facilitating HCC cell proliferation, migration and invasion[116]. By contrast, tumor suppressive lncRNAs that act as ceRNAs can exert their biological function by inactivating the PI3K/Akt pathway in HCC. For instance,TCL6, a tumor suppressive lncRNA, upregulates PTEN expression by sponging miR-106a-5p to suppress the PI3K/AKT pathway, thereby inhibiting HCC cell proliferation, migration, and invasion[117]. Intriguingly, several oncogenic lncRNAs that act as ceRNAs have been reported to exert their biological functions by activating the PI3K/AKT/mammalian rapamycin (mTOR) or PI3K/AKT/FoxO3a pathway in HCC. For example, the lncRNACDKN2B-AS1, an oncogenic lncRNA, enhances nucleosome assembly protein 1 like 1 expression by acting as a ceRNA of let-7c-5p, thus activating the PI3K/AKT/mTOR pathway to promote HCC cell growth and metastasis[118]. In addition, the lncRNAGAS6-AS2activates the PI3K/AKT /FoxO3a pathway by upregulating OTU domain-containing ubiquitin aldehydebinding protein 1 expression by sponging miR-493-5p, which promotes HCC cell proliferation, migration, and invasion[119]. In short, the lncRNA-miRNA-PI3K/AKT, PI3K/AKT/mTOR or PI3K-AKT-FoxO3a regulatory network is expected to be a potential therapeutic target for the treatment of HCC patients.
NF-κB pathway:Numerous studies have shown that abnormal activation of the NF-kB pathway is related to HCC growth, EMT, and invasion[120-122]. Existing evidence suggests that lncRNAs can act as miRNA sponges and exert their biological function by mediating the NF-κB pathway in HCC, among which oncogenic lncRNAs that act as ceRNAs can exert their biological function by activating the NF-κB pathway in HCC. For example, in the NF-κB pathway, mixed-lineage kinase (MLK) 3 contributes to cancer migration, invasion, and metastasis[123,124], and the lncRNASNHG12enhances MLK3 expression by competitively sponging miR-199a/b-5p, thereby activating the NF-κB pathway to promote HCC proliferation and tumorigenicity[125]. By contrast, a tumor suppressive lncRNA that acts as a ceRNA can exert its biological function by inactivating the NF-κB pathway in HCC. For example, CYLD, a tumor suppressor, can negatively regulate the NF-κB pathway in HCC[126], and the lncRNACASC2, a tumor suppressive lncRNA, suppresses the NF-κB pathway by enhancing CYLD expression by sponging miR-362-5p, thereby inhibiting HCC cell migration and invasion[127,128]. These findings indicate that the lncRNA-miRNA-NF-κB pathway network may serve as a therapeutic target for patients with HCC.
Chemoresistance and radioresistance
Although current chemotherapy and radiotherapy regimens can prolong the survival of HCC patients, tumor recurrence and metastasis due to chemoresistance and radioresistance lead to unsatisfactory long-term efficacy. The underlying mechanisms of therapeutic resistance in HCC are still unclear, and the exploration of such mechanisms will help improve the current treatment of HCC. Emerging evidence suggests that lncRNAs play a critical role in mediating chemoresistance and radioresistance by acting as ceRNAs of miRNAs in HCC.
Currently, the lncRNA-mediated ceRNA network has been proven to mediate HCC resistance to chemotherapy drugs, including sorafenib, oxaliplatin, cisplatin, and 5-fluorouracil (5-FU). Exploration of the resistance mechanisms to chemotherapy drugs in the treatment of HCC will provide new insights into overcoming chemoresistance.
Sorafenib has been approved for treating advanced HCC; however, the emergence of sorafenib resistance has affected the efficacy of HCC treatment. Existing studies have shown that the lncRNA-mediated ceRNA network is responsible for HCC resistance to sorafenib. Specifically, oncogenic lncRNAs that act as ceRNAs can enhance HCC resistance to sorafenib. For example, activation of the c-Met-Akt pathway can promote sorafenib resistance in HCC cells[129], and the lncRNANEAT1activates the c-Met-Akt pathway by sponging miR-335 to enhance sorafenib resistance in HCC cells[130]. In addition, recent studies have suggested that the abnormal activation or expression of forkhead box M1 (FoxM1) contributes to chemotherapy resistance in various cancer cells[131,132]. In HCC cells, FoxM1 knockout sensitizes drugresistant HCC cells to sorafenib[133], and the lncRNALINC-RORincreases sorafenib resistance in HCC cells by elevating FOXM1 expression by sponging miR-876-5p[134]. Phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3), a regulatory subunit of PI3K, activates the PI3K/AKT pathway to enhance the resistance of HCC cells to sorafenib-induced apoptosis[135], and the lncRNALINC00160acts as a miR-132 sponge to promote sorafenib resistance by increasing PIK3R3 expression in HCC cells[136]. Transmembrane protein 9 (TMEM9) plays a vital role in HCC cell growth[137], and the lncRNAFOXD2-AS1upregulates TMEM9 expression by sponging miR-150-5p to facilitate the resistance of HCC cells to sorafenib[138]. The tetraspanin protein CD151 has been shown to attenuate drug-induced apoptosis in cancer cell lines[139], and the lncRNASNHG3enhances CD151 expression by acting as a ceRNA for miR-128 to promote HCC resistance to sorafenib[140]. Additionally, the lncRNASNHG16is upregulated in sorafenib-resistant HCC cells, andSNHG16increases sorafenib resistance partly by competitively sponging miR-140-5p[141].
Oxaliplatin has been approved for the treatment of patients with locally advanced and metastatic HCC who are not eligible for surgical resection or local treatment; however, oxaliplatin resistance affects the efficacy of HCC treatment. The lncRNAmediated ceRNA network has been confirmed to modulate oxaliplatin resistance in HCC. In particular, oncogenic lncRNAs that act as ceRNAs can enhance HCC resistance to oxaliplatin. For instance,HULCcan upregulate the expression of the ubiquitin-specific peptidase 22 (USP22) protein by suppressing miR-6825-5p, miR-6845-5p, and miR-6886-3p at the epigenetic or transcriptional level in HCC cells; USP22 enhances theHULC-induced deubiquitination of Sirt1 and stabilizes it, and Sirt1 stability induces the autophagy of HCC cells, thus increasing the resistance of HCC cells to oxaliplatin[142]. Multidrug resistance-associated protein 1 (ABCC1) is indicative of chemotherapy resistance[143], and the lncRNANR2F1-AS1elevates ABCC1 expression by sponging miR-363 to enhance oxaliplatin resistance in HCC cells[144]. Similarly, the lncRNAKCNQ1OT1increases oxaliplatin resistance in HCC cells by sponging miR-7-5p to elevate ABCC1 expression[145].
The antitumor efficacy of cisplatin in the treatment of advanced HCC patients is unsatisfactory due to drug resistance. The lncRNA-mediated ceRNA network has been demonstrated to modulate cisplatin resistance in HCC, among which oncogenic lncRNAs that act as ceRNAs can enhance HCC resistance to cisplatin. For example, nuclear factor erythroid-2-related factor 2 (Nrf2) is upregulated in HepG2/cisplatin cells and mediates the chemoresistance of HCC cells to cisplatin[146]. The lncRNANRALincreases Nrf2 expression by sponging miR-340-5p, thereby facilitating cisplatin resistance in HCC cells[147]. Melanoma-associated antigen A3 (MAGEA3) enhances chemoresistance to cisplatin in HepG2 cells, and the lncRNALINC01234enhances MAGEA3 expression by sponging miR-31-5p to promote cisplatin resistance in HCC[148]. The lncRNASNHG16enhances cisplatin resistance in HCC cells by sponging let-7b-5p[149]. Conversely, tumor suppressive lncRNAs that act as ceRNAs can reduce HCC resistance to cisplatin. For example, the overexpression ofCASC2, a tumor suppressor lncRNA, strengthens cisplatin sensitivity in HCC cells by sponging miR-222[150]. In addition, the overexpression ofGAS5, another tumor suppressor lncRNA, enhances the sensitivity of HCC cells to cisplatin by sponging miR-222[151].
The inhibitory efficacy of 5-FU on HCC cells is limited by chemical resistance. Emerging evidence indicates that the lncRNA-mediated ceRNA network is correlated with 5-FU resistance in HCC cells, among which oncogenic lncRNAs that act as ceRNAs can enhance HCC resistance to 5-FU. For example, the lncRNACRNDEacts as a ceRNA of miR-33a in HCC to enhance HMGA2 expression, thereby promoting chemoresistance to 5-FU in HCC cells[152]. HIHIF-2α is related to the resistance of HCC cells to doxorubicin and sorafenib[153,154], and the lncRNAMALAT1acts as a ceRNA to increase HIF-2α expression by competitively sponging miR-216b, leading to the enhanced chemoresistance of HCC cells to 5-FU[155]. In contrast, tumor suppressive lncRNAs that act as ceRNAs can reduce HCC resistance to 5-FU. For example, Kelchlike ECH-associated protein 1 (Keap1) inactivation enhances the resistance of HCC cells to chemotherapy drugs such as sorafenib[156,157], and the overexpression ofKRAL, a tumor suppressive lncRNA, enhances Keap1 expression by functioning as a ceRNA for miR-141 to reverse the resistance to 5-FU in HCC cell lines[158].
Currently, an oncogenic lncRNA-mediated ceRNA network has been demonstrated to enhance HCC resistance to radiation therapy. For instance, AZD1775, an inhibitor of WEE1, has been reported to sensitize HCC cells to radiation[159], suggesting that WEE1 can enhance radioresistance in HCC. The lncRNANEAT1_2upregulates WEE1 expression by acting as a ceRNA for miR-101-3p to reduce the radiosensitivity of HCC[160]. A RING-type ubiquitin ligase E3 (RAD18), an E3 ubiquitin-linked enzyme, can induce radiation resistance in glioma cells[161,162]. The lncRNALINC-RORcompetes with sponge miR-145 to increase RAD18 expression, thereby enhancing the radiation resistance of HCC cells[163]. Forkhead box protein P1 (FOXP1), a transcription factor, attenuates radioresistance in cervical cancer[164]. The lncRNALINC00473promotes radioresistance in HCC by increasing FOXP1 expression by sponging miR-345-5p[165]. These findings suggest that the lncRNA-mediated ceRNA network may provide new clues for overcoming radioresistance in HCC.
PROBLEMS AND PERSPECTIVES
The lncRNA-mediated ceRNA regulatory network provides a new mode of posttranscriptional regulation and plays a critical role in the initiation and progression of HCC. Nevertheless, investigations into the detailed mechanism of the ceRNA network and its relationship with HCC are still in the preliminary stage. Although there are increasing reports about lncRNAs as ceRNAs in HCC, several fundamental problems facing these studies need to be addressed. First, information on the roles of lncRNAs that act as ceRNAs in current studies is derived from overexpression and/or knockout experiments, and only when the abundance of lncRNAs is remarkably high can lncRNAs act as ceRNAs. As a result, the abundance of artificially controlled lncRNAs often far exceeds the abundance range of any endogenous lncRNA. Therefore, it is urgent to verify whether the lncRNA-mediated ceRNA network has the same effects under normal cellular conditions. Second, most of the current research on ceRNAs is still in the prediction stage of bioinformatics, and most studies lack biological validation; the regulatory relationships in the ceRNA network need to be effectively verified. Third, methodologically, there are few predictive tools available; most miRNA-mRNA predictions focus only on the binding of a miRNA with its target in the 3’-UTR. However, this is not always the case; miRNAs can also target the 5'-untranslated region and coding sequences of mRNAs[166-170]. Thus, provided that the prediction of a ceRNA is not limited to the 3’-UTR of its mRNA, the range of predicted ceRNAs should be improved. Fourth, one miRNA generally interacts with one target mRNA. However, some miRNAs may modulate many target mRNAs, andvice versa[167,171]. Thus, it is necessary to model the effect of ceRNAs in real scenarios. Fifth, the ceRNA hypothesis maintains that a miRNA is stably expressed; in fact, intracellular miRNA expression is dynamic, which inevitably influences the effectiveness of ceRNAs. Sixth, although ceRNA prediction methods are constantly updated, the current prediction algorithms cannot fully encompass several factors affecting ceRNA susceptibility (quantity and characteristics of MREs, miRNA/mRNA abundance, and subcellular location of RNAs)[172,173]. Therefore, the prediction methods and experimental methods still need to be further updated and improved. Seventh, the initiation and progression of HCC are a complex event, and it is unclear whether other mechanisms interact with lncRNAs acting as ceRNAs in HCC cells. Addressing the above problems will enable a better understanding of the lncRNA-mediated ceRNA network that can be used to more effectively diagnose and treat HCC.
Given the critical roles and the complex interactions among lncRNA-mediated ceRNA regulatory networks in HCC, future investigations with validation in large sample sizes and the exploration of in-depth molecular mechanisms are needed to probe the HCC-specific lncRNA-mediated ceRNA axis, which should identify new diagnostic and prognostic markers of HCC and provide promising targets for the treatment of patients with HCC.
 World Journal of Gastroenterology2020年29期
World Journal of Gastroenterology2020年29期
- World Journal of Gastroenterology的其它文章
- Rno_circ_0005139 regulates apoptosis by targeting Wnt5a in rat anorectal malformations
- Gadoxetic acid magnetic-enhanced resonance imaging in the diagnosis of cholangiocarcinoma
- Impact of mobile health and medical applications on clinical practice in gastroenterology
- Multifocal gastrointestinal epithelioid angiosarcomas diagnosed by endoscopic mucosal resection: A case report
- Role of nutritional status and nutritional support in outcome of hepatitis B virus-associated acute-onchronic liver failure
- Dysregulation of microRNA in cholangiocarcinoma identified through a meta-analysis of microRNA profiling
