Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins?
Alexandre Loktionov
Abstract Colorectal cancer (CRC) is a global problem affecting millions of people worldwide. This disease is unique because of its slow progress that makes it preventable and often curable. CRC symptoms usually emerge only at advanced stages of the disease, consequently its early detection can be achieved only through active population screening, which markedly reduces mortality due to this cancer. CRC screening tests that employ non-invasively detectable biomarkers are currently being actively developed and, in most cases, samples of either stool or blood are used. However, alternative biological substances that can be collected non-invasively (colorectal mucus, urine, saliva, exhaled air) have now emerged as new sources of diagnostic biomarkers. The main categories of currently explored CRC biomarkers are: (1) Proteins (comprising widely used haemoglobin); (2) DNA (including mutations and methylation markers); (3) RNA(in particular microRNAs); (4) Low molecular weight metabolites (comprising volatile organic compounds) detectable by metabolomic techniques; and (5) Shifts in gut microbiome composition. Numerous tests for early CRC detection employing such non-invasive biomarkers have been proposed and clinically studied. While some of these studies generated promising early results, very few of the proposed tests have been transformed into clinically validated diagnostic/screening techniques. Such DNA-based tests as Food and Drug Administration-approved multitarget stool test (marketed as Cologuard?) or blood test for methylated septin 9 (marketed as Epi proColon? 2.0 CE) show good diagnostic performance but remain too expensive and technically complex to become effective CRC screening tools. It can be concluded that, despite its deficiencies, the protein (haemoglobin) detection-based faecal immunochemical test (FIT) today presents the most cost-effective option for non-invasive CRC screening. The combination of non-invasive FIT and confirmatory invasive colonoscopy is the current strategy of choice for CRC screening. However,continuing intense research in the area promises the emergence of new superior non-invasive CRC screening tests that will allow the development of improved disease prevention strategies.
Key words: Colorectal cancer screening; Biomarkers; Non-invasive testing; Stool;Colorectal mucus; Blood
INTRODUCTION
Colorectal cancer (CRC) is currently the third most frequently diagnosed cancer worldwide. The global incidence for 2018 is estimated at 1801000 new cases, and the number of CRC-related deaths for this period is 861700[1]. Although the highest CRC incidence continues to be observed in economically developed Western countries, it is now rapidly increasing in other parts of the world[2]. Sporadic CRC development can take decades and is in most cases characterised by a slow progression from aberrant crypt formation in the colonic mucosa to benign polyps that may give rise to early cancer, then gradually evolving to invasive and metastasising advanced neoplasms(Figure 1)[2-4]. These pathogenetic features make CRC one of the most preventable and often curable malignancies. However, disease curability entirely depends on its early detection, which is not straightforward as clinical symptoms usually emerge only when CRC is already advanced. The latter factor warrants the necessity of active population screening for CRC, and it has been well proven that screening saves lives[2].
Full colonoscopy is regarded as the gold standard diagnostic technique for colorectal tumour detection[5], and it has become a very popular method for primary CRC screening[6-8]in the United States. One apparent reason for this trend is that diagnostic colonoscopy is usually combined with the simultaneous removal of detected polyps and functions as both a diagnostic and preventive procedure clearly reducing mortality from CRC[9]. Nonetheless, colonoscopy is an expensive and invasive technique that requires unpleasant bowel preparation and occasionally causes serious complications[10]. Moreover, its sensitivity is not perfect, with polyps sometimes missed[11], the latter problem often depending on the operator’s skills[12].Although colonoscopy as the final (confirmatory) diagnostic step is undisputable, its use in primary CRC screening remains questionable as indiscriminate application of this method inevitably results in frequent negative outcomes and a large health economic burden[13]. In theory, the global introduction of non-invasive tests employing biomarker analysis to select patients that really require endoscopy could dramatically reduce the numbers of unnecessary colonoscopies. Unfortunately, none of the existing non-invasive tests successfully combine high diagnostic sensitivity and specificity with technical simplicity and low cost, the key characteristics of an ideal screening modality. This paper provides a brief overview of the current state of the area encompassing biomarker-based non-invasive tests for CRC detection.
SOURCES OF MATERIAL FOR NON-INVASIVE CRC BIOMARKER TESTING
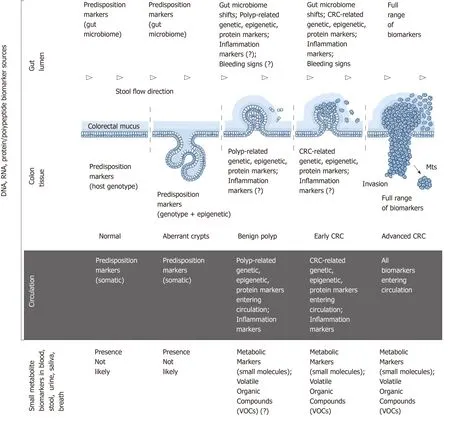
Figure 1 Colorectal cancer pathogenesis and sources of potential diagnostic biomarkers at different stages of colorectal cancer development. CRC:Colorectal cancer.
CRC development is an extraordinarily complex process driven by multiple genetic,epigenetic, metabolic and immune alterations at the host level and influenced by numerous environmental factors[4,14,15]. Despite intense research, precise mechanisms of CRC development remain largely obscure[4,14,15]. Genome-targeting investigations,especially genome-wide association studies, have revealed a highly complex pathogenetic landscape comprising multiple alternative cascades of molecular events that may eventually result in cancer[4,16]. This complexity leads some investigators to a hardly satisfactory conclusion that “each patient’s CRC is genetically and epigenetically unique”[4]. Nevertheless, colorectal tumours frequently have common molecular patterns that are diagnostically relevant and will be considered below.
The series of morphological events accompanying CRC development is presented in Figure 1. This sequence involves numerous associations with various types of biomolecules that can be characterised as biomarkers. The ideal biomarkers for CRC can be defined as substances that satisfy the following criteria: “(1) Are measured easily and inexpensively to identify a patient’s cancer; (2) Identify a patient’s prognosis to improve treatment outcome; and (3) Predict a patient’s response to a specific treatment”[15]. This paper is focused only on the first category,i.e., diagnostic biomarkers of CRC that can be sampled and tested non-invasively.
Figure 1 outlines the main sources of CRC biomarkers in relation to disease stages.From the morphological point of view, it is obvious that (1) colon tissue; (2) gut lumen; (3) blood/lymph circulation are the main sources of CRC-associated DNA,RNA and protein/polypeptide biomarkers associated with the host; (4) moreover,specific pattern shifts in small metabolite molecules derived from CRC-affected metabolic pathways constitute an additional group of post-metabolic markers that can be analysed by metabolomics techniques[17,18]; and (5) CRC-associated gut microbiome changes[19]deserve to be considered as a separate category of diagnostic markers of non-human origin.
Normal and neoplastic colon tissue
Colonic epithelium is the site of neoplastic growth initiation. After that CRC progresses within the colonic wall until advanced stages of the disease, hence premalignant and malignant colon tissues are certainly the richest biomarker sources[4].However, invasive biopsies are required for sampling tissue. Therefore, CRC markers detectable in tissue samples are not discussed here.
Gut lumen
Colonic epithelium is the key element of the gastrointestinal barrier between host tissues and microbiota-rich colon contents. Until recently it was presumed that all host cells exfoliated or migrated from the surface of the colonic epithelium were immediately incorporated in the faecal matter. According to this simplistic notion, it seemed to be logical that analysing naturally excreted stool samples constitutes the only perfectly non-invasive approach to investigating CRC biomarkers. It should,however, be stressed that stool is a complex mixture of microbiota-dominated faecal matter and occasional fragments of colorectal mucus secreted by goblet cells of the colonic epithelium. While the prevailing faecal component of stool entirely belongs to the environment, colorectal mucus is host-derived. The two-layered structure and functional significance of the mucus overlaying colonic epithelium have been elucidated only during the last decade[20,21], and it is now clear that colorectal mucus rather than faecal matter is the main receptacle of all cells and biomolecules released from either normal or malignant epithelium[22,23]. Intrarectal collection of colorectal mucus had demonstrated high informativeness of this substance[22,23], which was shown to accept CRC-generated malignant colonocytes exfoliated from tumour surface and transport them distally alongside stool flow without incorporating them into faeces (Figure 1)[20,23]. Βiomarker-rich colorectal mucus essentially serves as a border between well oxygenated colonic epithelium and anaerobic gut lumen. Our group has recently developed a simple technique for non-invasive sampling of this mucus[24-26], the analysis of which may constitute a very convenient alternative to stoolbased tests.
Blood/lymph circulation
Βlood-derived biomarker analysis is another area of significant interest in the context of CRC detection since blood collection is regarded as a practically non-invasive procedure. It is evident that a wide range of CRC-associated biomarkers can be detected in the circulating blood and lymph of patients with these malignancies, but lymph collection cannot be performed with minimal invasiveness. For this reason,only biomarkers measurable in blood will be discussed below. In the modern literature the term “l(fā)iquid biopsy” is often applied to this group of biomarker-based techniques[27]. Nevertheless, despite the easiness of blood sampling and the availability of numerous analytical techniques for biomarker detection in human plasma or serum, the presence of cancer biomarkers in blood may or may not be associated with CRC. Malignancies of other sites should always be excluded if this approach is considered for CRC screening.
Post-metabolic biomarkers
The use of metabolomics for revealing CRC-specific changes in patterns of low molecular weight metabolites has recently become another area of active exploration[28]. This new approach can potentially employ a wider range of biological samples comprising blood, stool, colorectal mucus, urine, saliva and exhaled breath,thus bringing about additional diagnostic options.
Gut microbiome changes associated with CRC
Recent research has revealed that specific chang es in gut microbiome composition may be associated with the development of CRC[19]. In this context stool samples are usually investigated quantitatively for the presence of particular types of bacteria.
The limited choice of sample sources for non-invasive testing creates obvious problems. Collecting gut-derived samples looks preferable, but stool samples, albeit containing cells and molecules originating from the colonic mucosa (i.e., colorectal mucus fragments), are usually dominated by the presence of abundant microbiotarich faecal matter that often interferes with analytical procedures employed for hostrelated biomarker detection. A recently described analysis of non-invasively collected colorectal mucus presents a very interesting alternative; however, this approach is new and requires further testing. On the other hand, blood collection is very straightforward and easy to standardise, but molecular changes detectable in blood(or plasma/serum) samples are not necessarily gut-specific. Finally, although the use of easily collectable materials (urine, saliva or exhaled air) is extremely attractive, the presence of CRC-specific biomarkers in such samples remains to be adequately explored. The sources of biological material characterised above may contain several types of diagnostic biomarkers that are discussed in the next section.
BIOMARKERS ASSOCIATED WITH CRC DEVELOPMENT
The story of non-invasively detectable CRC markers started due to a 1967 publication by Greegor, describing his observation of the frequent presence of occult blood in stool samples collected from patients with CRC[29]. That important discovery resulted in the development and prolonged use of the haemoglobin-recognising faecal occult blood test (FOΒT) as the only non-invasive test for CRC detection. The situation had changed considerably in 1992, when a publication by Sidranskyet al[30]described Κ-ras gene mutation detection in stool samples obtained from CRC patients and shifted the focus of attention to molecular markers. The area of CRC biomarker research has since exponentially expanded with thousands of papers published, but many initially promising findings failed to transform into clinically relevant diagnostic approaches.The purpose of this paper is to briefly outline the present status of non-invasive biomarkers proposed for detecting asymptomatic CRC. Only the most impressive and clinically relevant observations related to the main groups of these biomarkers(proteins/polypeptides, DNA, RNA, small metabolites, microbiome changes) are highlighted in the text below. However, numerous other markers that demonstrated promise in the context of CRC detection are presented in comprehensive Tables 1, 2, 3,4 and 5. As it was impossible to cover all relevant studies, restrictions had to be applied when the Tables were prepared. Publications describing very small studies or reporting negative results were omitted. Likewise, only papers related to CRC, but not adenoma detection, were included since in most cases diagnostic sensitivity of biomarker tests for adenomatous polyps correlates with that for CRC. In addition, the necessity of non-invasive detection of colorectal polyps is still a debatable question, as the proportion of adenomas likely to progress to malignancy is relatively small,whereas the vast majority of these lesions (especially small polyps) never give rise to CRC[134,135].
Protein markers
Protein biomarkers considered in CRC early detection and screening are listed in Table 1. Historically, the use of haemoglobin detection in stool for non-invasive CRC detection can be regarded as the most popular approach in terms of population screening. Indeed, the traditional guaiac FOΒT was almost exclusively employed for this purpose for several decades, and was attractive due to its simplicity and low cost.Although this test has insufficient sensitivity, it can be credited for saving many human lives[2,136,137]. Nevertheless, the outdated FOΒT is now being replaced by the faecal immunochemical test (FIT) characterised by a much higher sensitivity. In a recent comprehensive review on FIT, Gieset al[31]discussed numerous studies of varying sizes and reported sensitivities between 66% and 74% and specificity levels between 84% and 95% when numbers of analysed CRC cases and controls were over 50. Table 1 also shows that M2-pyruvate kinase (M2-PΚ) is a relatively well-studied stool marker of CRC[32,33]; however, FIT performs better and remains considerably more popular. Other stool tests, including metalloproteinase 9 (MMP9)[34]and multimarker protein panels (see Table 1) have been investigated, but these tests have not been clinically accepted so far. It is also intriguing that in a recent small study, our group compared 24 protein biomarkers in non-invasively collected samples of colorectal mucus and concluded that haemoglobin, tissue inhibitor of metalloproteinase 1, M2-PΚ, peptidyl arginine deiminase 1, C-reactive protein and MMP9 could reliably detect CRC[138].
Βlood (or plasma/serum) testing for CRC-associated proteins has been employed by many research groups (Table 1), but most of those studies produced relatively modest results. Among single protein markers detectable in the serum only CA11-19 marker protein[36], cysteine-rich 61 protein of the CCN family (Cyr 61)[38], Β6-integrin[39]and trefoil factor 3 (TFF3)[36]can be regarded as promising. A number of protein panels were also examined; however, analysing multiple proteins is usually more technically complex and expensive. Impressive test sensitivity and specificity values(98.7% and 94.8%, respectively) were reported for combined testing for lectins DCSIGN and DC-SIGNR by Jianget al[42]in 2014, but these results remain to be confirmed in larger studies. Although blood collection is simple and easy to standardise, proteinbiomarkers of CRC present in stool or colorectal mucus currently look more diagnostically reliable than those detectable in blood.

Table 1 Non-invasive protein (including cytokine) biomarkers used for colorectal cancer detection
An additional advantage of using protein biomarkers for CRC detection is defined by the fact that their immunochemical detection can be easily presented as point of care (POC) tests, which are already available for FIT[139].
DNA and mRNA markers
This sub-section briefly discusses studies on CRC detection using DNA and mRNA markers that are listed in Table 2.
Gene mutations, especially those ofK-RasandAPCgenes, were the first CRCassociated genetic markers assessed with the purpose of developing new non-invasive modalities for CRC early detection and screening. Regrettably, it soon became clearthat using gene mutations alone does not achieve satisfactory levels of diagnostic sensitivity. One demonstrative study evaluating this approach in a representative colonoscopy screening group concluded that the sensitivity of a panel comprising 21 DNA alterations (point mutations inK-ras,APCandp53genes, microsatellite instability marker ΒAT-26 deletions and long DNA assay) was only slightly above 50%[46].
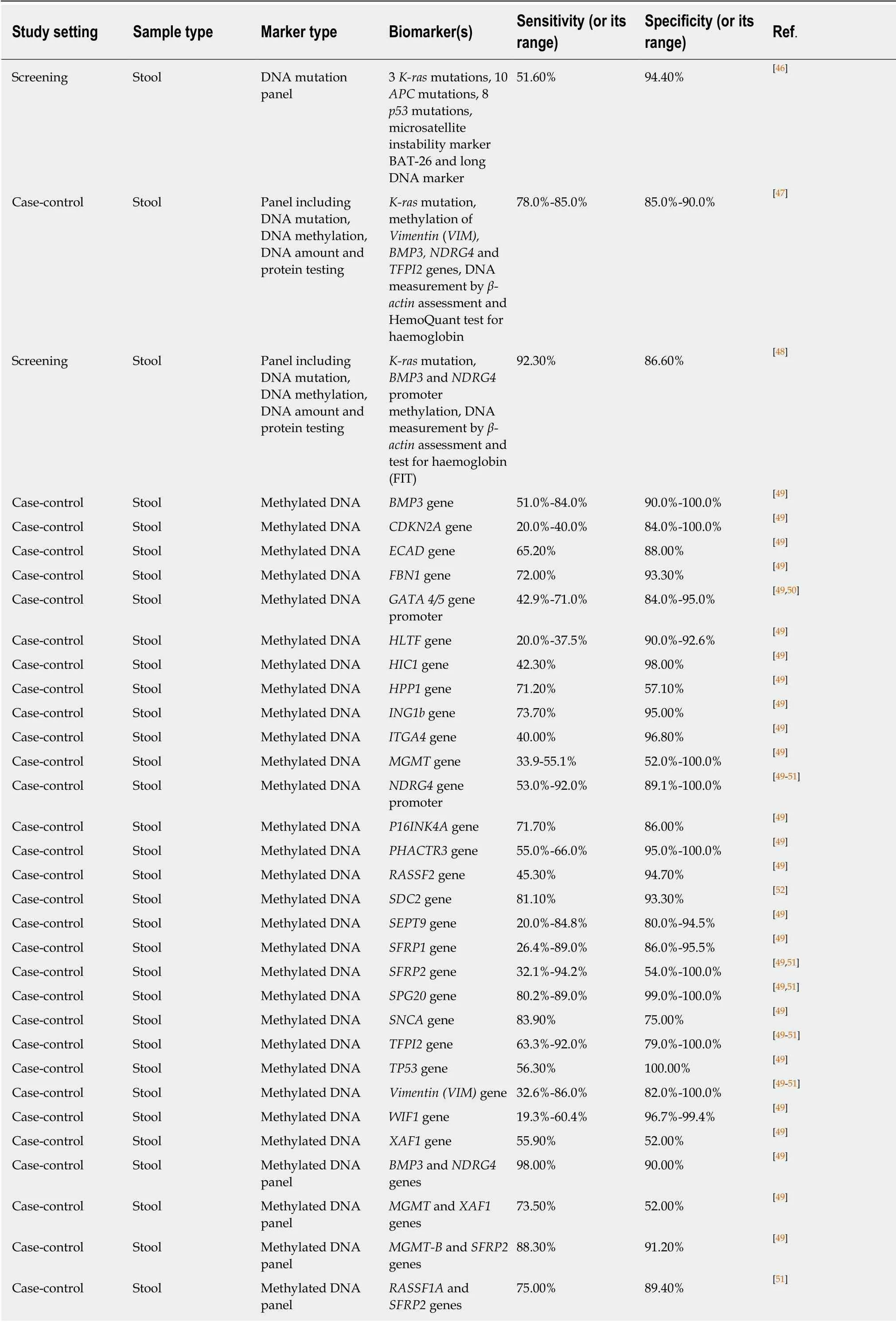
Table 2 Non-invasive DNA, messenger RNA and long non-coding RNA biomarkers used for colorectal cancer detection
84.30% 93.30% [53]Case-control Stool Methylated DNA panel SNCA and FNB1 genes Case-control Stool Methylated DNA panel Vimentin (VIM) and SFRP2 genes 92.50% 91.20% [53]Case-control Stool Methylated DNA panel AGTR1, WNT2 and SLIT2 genes 74.0%-78.0% 88.0%-89.0% [49,50]Case-control Stool Methylated DNA panel ECAD, MGMT and P16INK4A genes 72.00% 88.00% [49]Case-control Stool Methylated DNA panel ITGA4, SFRP2 and P16INK4A genes 70.00% 96.80% [49]Case-control Stool Methylated DNA panel MGMT, CDKN2A and hMTH1 genes 55.00% 63.00% [49]Case-control Stool Methylated DNA panel MGMT, MLH1 and Vimentin (VIM)genes 75.00% 86.50% [49,51]Case-control Stool Methylated DNA panel SFRP2, HPP1 and MGMT genes 93.70% 77.10% [49]Case-control Stool Methylated DNA panel WIF-1, ALX-4 and Vimentin (VIM)genes 25.00% 98.00% [49]Case-control Stool Methylated DNA panel Vimentin (VIM),OMSR and TFPI2 genes 86.70% 87.60% [49]Case-control Stool Methylated DNA panel SFRP2, GATA4/5,NRDG4 and Vimentin (VIM)genes 96.40% 65.00% [49]Case-control Stool Human DNA content Total human DNA content 66.00% 89.80% [54]Case-control Βowel Lavage Fluid Methylated DNA panel miR-124-3,LOC386758 and SFRP1 genes 82.00% 79.00% [55]Case-control Intrarectally collected colorectal mucus Human DNA content Total human DNA content 60.40% 94.80% [56]Case-control Serum/plasma Methylated DNA ALX4 gene 23.0%-90.7% 72.5%-100.0% [57]Case-control Serum/plasma Methylated DNA APC gene 57.0%-86.5% 86.0%-92.1% [57]Case-control Plasma Methylated DNA CDH1 (E-cadherin)gene 60.00% 84.00% [55]Case-control Serum/plasma Methylated DNA SDC2 gene 87.0%-90.7% 72.5%-95.2% [36,57]Case-control Serum/plasma Methylated DNA SEPT9 gene 47.1-95.6% 81.0%-96.7% [36,57-62]Case-control Serum/plasma Methylated DNA SFRP2 gene 54.0%-69.4% 40.0%-98.7% [57,63]Case-control Plasma Methylated DNA THBD(Thrombomodulin)gene 70.70% 80.30% [51]Case-control Serum/plasma Methylated DNA TPEF gene 65.0%-81.0% 69.0%-90.0% [57]Case-control Serum/plasma Methylated DNA VIM (Vimentin) gene 59.0%-90.7% 72.5%-93.0% [57]Case-control Plasma Hypomethylated DNA LINE-1 transposable DNA element 65.80% 90.00% [36]Case-control Serum/plasma Methylated DNA panel IKFZ and BCAT1 genes 62.1%-95.0% 92.0%-95.0% [36,57]Case-control Serum Methylated DNA panel SEPT9 and SDC2 genes 86.50% 92.10% [64]Case-control Serum/plasma Methylated DNA panel APC, MGMT,RASSF2A and WIF-1 genes 86.50% 92.10% [57]Case-control Plasma Methylated DNA panel ALX4, BMP3,NPTX2, RARB,SDC2, SEPT9 and VIM genes 90.70% 72.50% [63]Case-control Serum ALU115 DNA content Free ALU115 DNA content 69.20% 99.10% [36]Case-control Serum DNA integrity ALU247/115 DNA integrity index 73.10% 97.30% [36]Case-control Serum Free DNA content ALU-based cell-free DNA 64.50% 98.90% [36]

FIT: Faecal immunochemical test; CRC: Colorectal cancer.
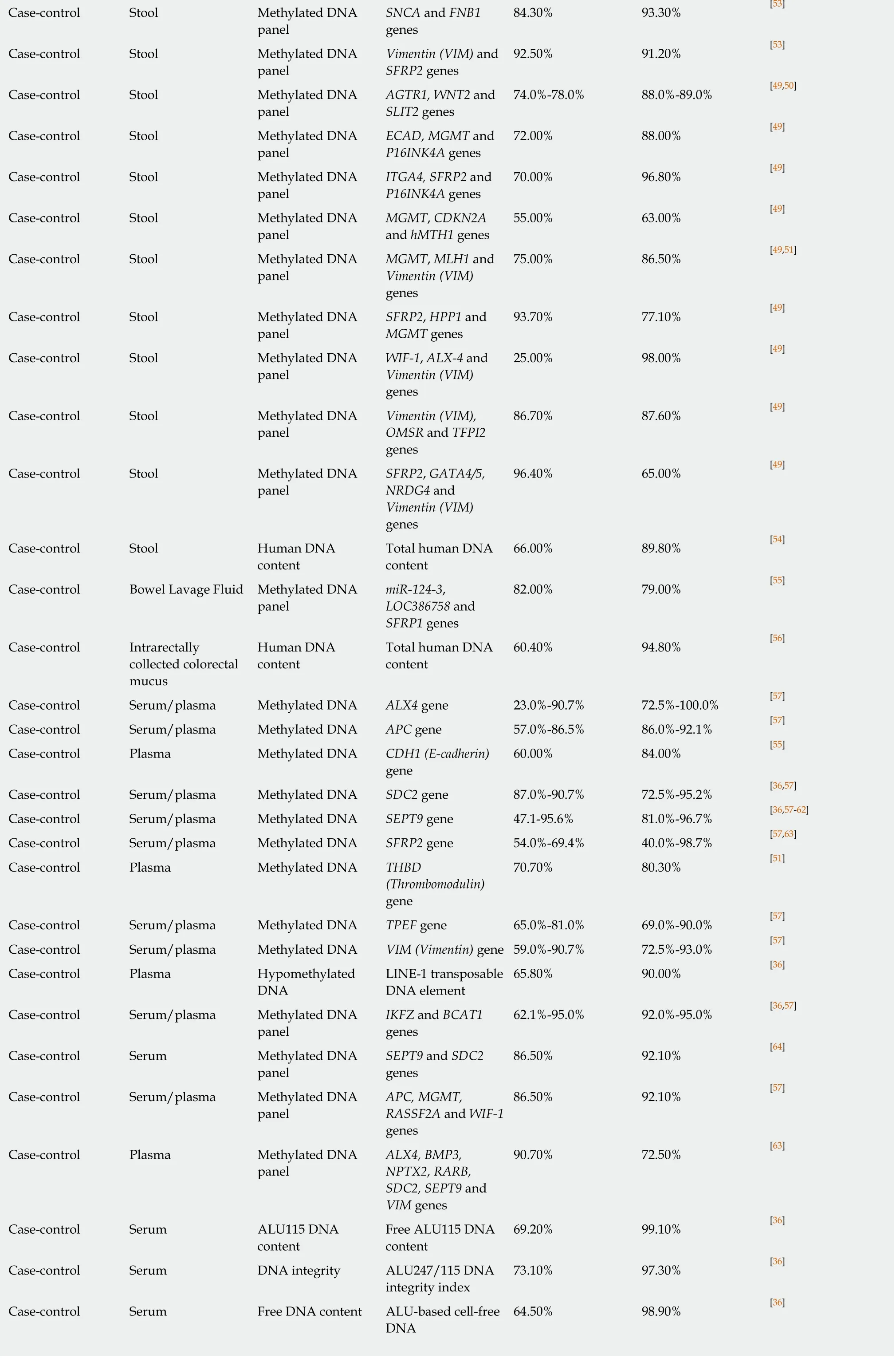
The relatively disappointing diagnostic performance of mutation-based assays stimulated the search for CRC-related epigenetic changes, in particular aberrant hypermethylation of CpG islands usually located in gene promoter regions[140]. Genespecific DNA methylation in stool was extensively investigated (Table 2), and several genes, includingBMP3,NDGR4,septin 9(SEPT9),SFRP2, SPG20, TFPI2andvimentin(VIM) were shown to have diagnostic sensitivities between 50% and 92% at specificities between 80% and 100% for CRC detection (see recent reviews by Liuet al[49], Lamet al[50]and Rasmussenet al[51]). However, the reproducibility of these results was often problematic, and attempts to combine multiple methylated genes in panels were undertaken to increase assay reliability. It is remarkable that high CRC detection sensitivity and specificity values could be achieved by combining methylation testing forBMP3andNDRG4[49]orVIMandSFRP2[53]genes, but these results need to be corroborated. The ColosureTMtest detecting methylatedVIMin stool was the first methylation-based commercial test for CRC[141]. This diagnostic product was marketed in the USA but has recently been replaced by a more efficient multimarker Cologuard?test considered later in this sub-section.
Table 2 demonstrates that in the context of CRC diagnostics, DNA methylation markers detectable in blood attract at least as much attention as similar markers in stool. Although investigations of different groups often produce conflicting results, it is now apparent thatSEPT9methylation detection is the best studied option amongst these blood tests[57]. This test has recently been commercialised and regulated for clinical application as Epi proColon?2.0 CE[142], but its use appears to be limited to opportunistic CRC screening[57]. Moreover, DNA methylation analysis in biological samples is relatively laborious (especially for multimarker panels) and difficult to present in POC format. These factors limit diagnostic potential of this approach. In addition, Table 2 shows that samples of stool, blood, bowel lavage fluid and colorectal mucus were also tested for total and ALU-based DNA quantification, DNA integrity assessment, examination of gene expression and long non-coding RNA expression.However, none of these assays could provide sufficiently high values for diagnostic sensitivity and specificity.
It is now becoming clear that tests involving DNA markers tend to perform better only when markers of different types are combined. Long-term research projects led by a United States company, Exact Sciences, allowed the design of a multitarget stool test that demonstrated high levels of sensitivity and specificity for CRC detection. An early version of this test that includedK-rasmutation, methylation ofVIM, BMP3,NDRG4andTFPI2genes, DNA measurement byβ-actinassessment and the HemoQuant test for haemoglobin achieved diagnostic sensitivity between 78% and 85% at specificity between 85% and 90% in a case-control study[47]. It is remarkable that this test performed significantly better when directly compared with the test for methylatedSEPT9in plasma (similar to Epi proColon)[143]. The multitarget test was then simplified, and its final version includes only determination ofK-rasmutation,BMP3andNDRG4promoter methylation, DNA measurement byβ-actinassessment and FIT. Screening application of this test in a large study produced CRC detection sensitivity of 92.3% at a specificity of 86.6%[48], which makes this assay the best among all available tests involving DNA markers. The test was approved by the United States Food and Drug Administration in 2014 and is now marketed as Cologuard?.However, this test, which can be regarded as an enhanced version of FIT, requires stool collection, remains technically complex, with a multistep analytical procedure required[144], and is very expensive at over $600.
MicroRNA markers
MicroRNAs (a sub-class of small non-coding RNA molecules) were discovered and characterised during the last decade of the XX century. Since that time, it was established that microRNAs are important regulators of gene expression intimately involved in the pathogenesis of many diseases including cancer[145]. As many of them are associated with the presence of colorectal tumours, it was suggested that microRNA determination in stool or blood samples may provide a new diagnostic modality for CRC early detection and screening[73]. MicroRNA variants investigated as potential CRC markers are listed in Table 3. Several published studies that used stool sample analysis highlight miR-21 as the best-studied marker of this type, but do not show outstanding sensitivity and specificity values[73]. MiR-451 and miR-223 detectable in stool produced high sensitivity and specificity values in a small study[75];however, these markers looked less impressive in other studies, when combined with other microRNAs[73,76]. It is impossible to exclude that these discrepancies may be associated with either technical problems or different ethnic composition of the studied patient groups since clinical studies providing material for microRNA analyses were performed mostly in East Asia.
Table 3 also indicates that microRNA markers of CRC were intensely investigated in blood. Hitherto most of these studies produced modest or inconsistent results.Again, miR-21 was assessed by many groups, and conflicting results were published.Although very high diagnostic sensitivity (96.6%) and specificity (97.8%) values were reported by Nget al[80]for miR-139-3p, which was shown to be downregulated in the serum of CRC patients, this finding remains to be confirmed. Combinations of microRNA markers detectable in plasma or serum were also tested as diagnostic panels. Among these panels (Table 3), combinations of downregulated miR-144-3p,miR-425-5p and miR-1260b[88]and upregulated miR-19a, miR-19b, miR-15b, miR-29a,miR-335 and miR-18a[90]demonstrated sensitivity and specificity levels exceeding 90%.
In addition, it should be noted that a recent small study has revealed that quantification of miR-21 in saliva samples resulted in CRC detection with 97%sensitivity and 91% specificity[93]. However, these highly intriguing results remain to be corroborated.
Although microRNAs constitute a group of promising CRC biomarkers, further research in this relatively new area is needed to establish clinically valid diagnostic techniques using these markers. The relative technical complexity of laboratory procedures used in microRNA analysis (RNA extraction, reverse transcription and qPCR analysis) and the necessity of careful assay optimisation and standardisation[146]should also be taken into account when the diagnostic potential of this interesting approach is considered.
Volatile organic compounds (VOC) and small metabolite biomarkers
Metabolomics is a new discipline that focuses on evaluating a wide variety of endogenous metabolites produced by the organism[17,18,28]. These metabolites can serve as late stage biomarkers of either normal physiological or pathophysiological events,and cancer metabolome is defined as the entire suite of low molecular weight (< 1500 Da) cancer-specific metabolites[17]. Interestingly, some of these metabolites are VOC-sthat are present in the gas phase of various excreted biological materials and can potentially be used for detecting malignancies including CRC[99]. The outcomes of metabolomic studies on CRC detection are summarised in Table 4. Remarkably, very impressive results (with CRC detection sensitivity reaching 97% at 99% specificity)were achieved by Sonodaet al[97], when dog scent judgment was applied to faeces and exhaled breath samples for discriminating between CRC patients and controls.Unfortunately, it is not realistic to expect that this natural phenomenon could constitute a reliable diagnostic tool. Hence, advanced Electronic Nose technologies are being developed and tested for CRC detection (Table 4) alongside widely used combinations of gas chromatography (GC) and mass spectrometry (MS)[18,94,99]. The latter approach, albeit regarded as the technical gold standard, is complex, costly and unsuitable for population screening. This point is especially important because most of the numerous studies applying metabolomic approaches to detecting CRC-related metabolites (non-VOC-s) in biological substances use various versions of MS (Table 4). Although some of the studies listed in Table 4 produced sensitivity and specificity values above 90% for CRC detection[102,109,113,116,125], cost and complexity issues remain major obstacles to the introduction of these assays into routine clinical practice. In this context, the use of electronic noses sensing CRC-associated VOC-s appears to be more promising, especially in view of CRC detection sensitivity and specificity both reaching 95% in a recent study by Zontaet al[98].
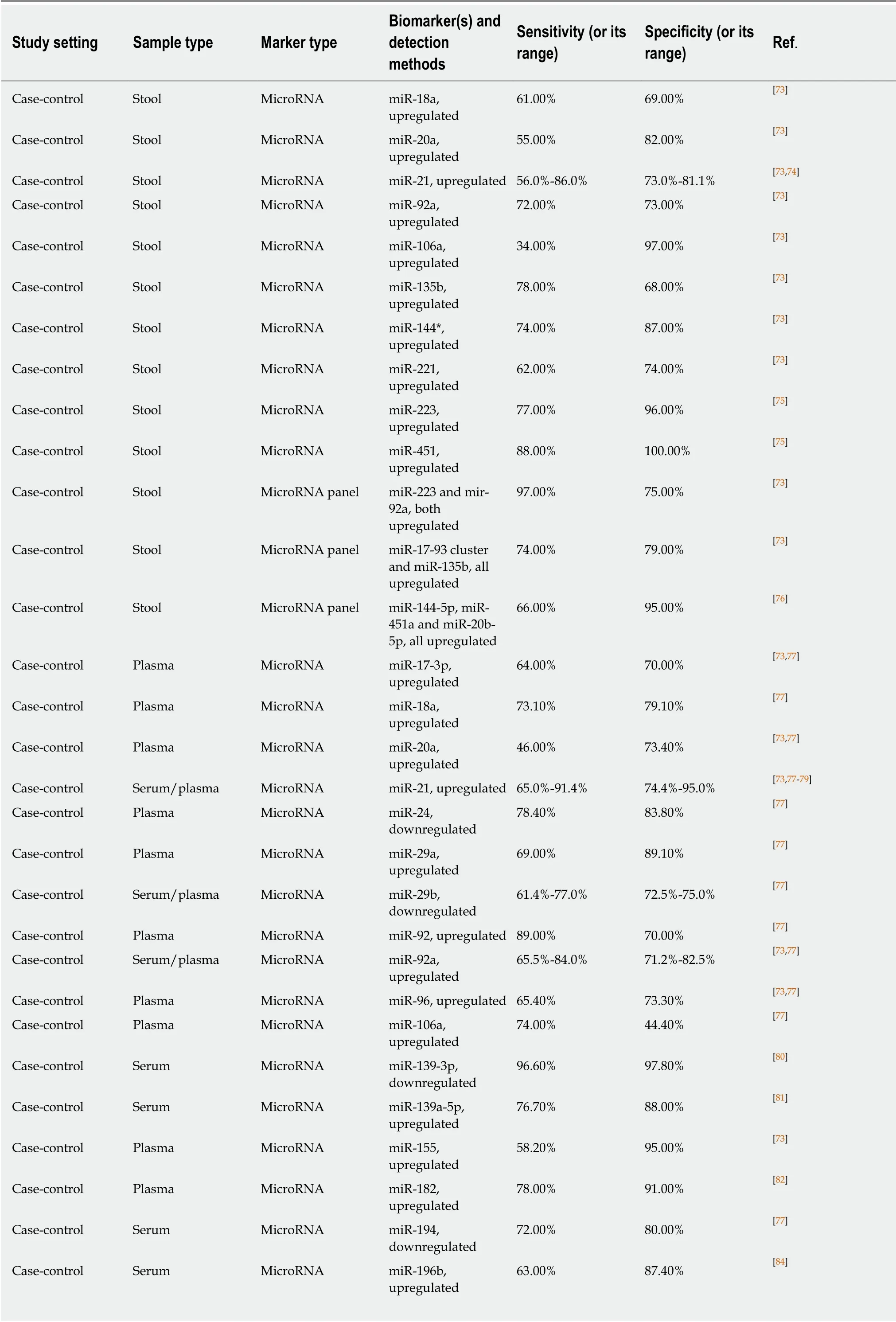
Table 3 Non-invasive microRNA biomarkers used for colorectal cancer detection

Case-control Plasma MicroRNA miR-200c,upregulated 64.10% 73.30% [77]Case-control Serum MicroRNA miR-210,upregulated 74.6%-88.6% 73.5%-90.1% [77,79]Case-control Plasma MicroRNA miR-221,upregulated 86.00% 41.00% [73,77]Case-control Plasma MicroRNA miR-320a,downregulated 92.80% 73.10% [77]Case-control Serum MicroRNA miR-338-5p,upregulated 76.30% 92.50% [84]Case-control Serum MicroRNA miR-372,upregulated 81.90% 73.30% [77]Case-control Serum MicroRNA miR-375,downregulated 76.90% 64.60% [77]Case-control Plasma MicroRNA miR-423-5p,downregulated 91.90% 70.80% [77]Case-control Plasma MicroRNA miR-506,upregulated 76.80% 60.70% [85]Case-control Plasma MicroRNA miR-601,downregulated 69.20% 72.40% [77]Case-control Plasma MicroRNA miR-760,downregulated 80.00% 72.40% [77]Case-control Serum MicroRNA miR-1290,upregulated 70.10% 91.20% [86]Case-control Plasma MicroRNA miR-4316,upregulated 76.80% 75.00% [85]Case-control Plasma MicroRNA panel miR-19a and miR-19b, both upregulated 78.60% 77.40% [77]Case-control Serum MicroRNA panel miR-21 and miR-92a,both upregulated 68.00% 91.20% [73,77]Case-control Plasma MicroRNA panel miR-29a and miR-92a, both upregulated 83.00% 84.70% [73,77]Case-control Plasma MicroRNA panel miR-200c and miR-18a, both upregulated 84.60% 75.60% [36,77]Case-control Plasma MicroRNA panel miR-223 and miR-92a, both upregulated 76.00% 71.00% [73]Case-control Plasma MicroRNA panel miR-320d,downregulated;miR-1290,upregulated 81.20% 90.70% [87]Case-control Plasma MicroRNA panel miR-431 and miR-139-p3, both upregulated 91.00% 57.00% [77]Case-control Plasma MicroRNA panel miR-601 and miR-760, both downregulated 83.30% 69.10% [73,77]Case-control Plasma MicroRNA panel miR-19a, miR-19b and miR-15b, all upregulated 78.60% 79.20% [77]Case-control Plasma MicroRNA panel miR-24, miR-320a and miR-423-5p, all downregulated 92.80% 70.80% [36,77]Case-control Plasma MicroRNA panel miR-144-3p, miR-425-5p and miR-1260b, all downregulated 93.80% 91.30% [88]Case-control Serum MicroRNA panel miR-145,downregulated;miR-106a and miR-17-3p, upregulated 78.50% 82.80% [73,77]Case-control Plasma MicroRNA panel miR-409-3p,upregulated; miR-7 and miR-93,downregulated 82.00% 89.00% [73,77]

Case-control Plasma MicroRNA panel miR-18a, miR-21,miR-22 and miR-25,all upregulated 67.00% 90.00% [89]Case-control Serum MicroRNA panel miR-23a-3p, miR-27a-3p, miR-142-5p and miR-376c-3p, all upregulated 89.00% 81% [36]Case-control Plasma MicroRNA panel miR-29a, miR-92a,upregulated; miR-601, miR-760,downregulated 83.30% 93.10% [77]Case-control Serum MicroRNA panel miR-21, miR-29,miR-92, miR-125,miR-223, all upregulated 84.70% 98.70% [78]Case-control Plasma MicroRNA panel miR-19a, miR-19b,miR-15b, miR-29a,miR-335, miR-18a,all upregulated 91.00% 90.00% [90]Case-control Plasma MicroRNA panel miR-21, let-7g,upregulated, mir-31,mir-92a, miR-181b,miR-203,downregulated 96.00% 81.00% [73]Case-control Plasma MicroRNA panel miR-103a-3p, miR-127-3p, miR-151a-5p,miR-17-5p, miR-181a-3p, miR-18a-5p,miR-18b-5p, all upregulated 76.90% 86.70% [91]Case-control Plasma Exosomal MicroRNA panel miR-27a, miR-130a,both upregulated 82.50% 75.00% [92]Case-control Saliva MicroRNA miR-21, upregulated 97.00% 91.00% [93]
Markers of CRC-associated changes in gut microbiome
The structure of the gastrointestinal tract engenders permanent interactions between its epithelial tissue and luminal microbiota, thus significant microbial impact in colorectal carcinogenesis appears to be likelier than in any other neoplasia. Steadily accumulating evidence indicates a pivotal role for the gut microbiome in influencing the development of CRC[19]. It is now believed that bacterial effects predisposing to CRC include impacts in gut surface barrier disruption, induction of colonic inflammation, direct genotoxic action against epithelial cells and dysbiosis leading to CRC-promoting shifts in gut microflora composition and the colonic microenvironment[19,147]. These advances prompted interest in evaluating gut microbiome shifts as possible diagnostic markers for CRC[148]. The results of several recent studies (presented in Table 5) show that alterations in gut microbiome composition can potentially serve as non-invasive diagnostic markers for this disease.

Table 4 Non-invasive volatile organic compounds and small metabolite biomarkers used for colorectal cancer detection

91.20% 93.50% [102]Case-control Stool Small metabolites Succinate – detected by PMRS Case-control Serum Aromatic carboxylic acids Βenzoic acid –detected by CE-time of flight (TOF) MS 89.00% 82.00% [103]Case-control Serum Fatty acids GTA-446 – detected by flow injection analysis MS 83.30% 84.80% [104]Case-control Plasma Amino acid metabolites L-kynurenine –detected by highperformance liquid chromatography(HPLC)85.20% 100.00% [105]Case-control Plasma Fatty acids Decanoic acid –detected by CETOFMS 87.80% 80.00% [106]Case-control Serum Multiple metabolites 38 metabolites detected by GC-MS 85.00% 86.00% [107]Case-control Serum Phospholipids(sphingomyelins and phosphatidylcholines)SM (34:1), PC (34:1),PC (34:2), PC (36:4),PC (36:2), PC (36:3) -detected by MS♂77.3%; ♀80.8% ♂92.4%; ♀85.9% [108]Case-control Serum Unsaturated free fatty acids (panel)C16:1, C18:3, C20:4,C22:6, all downregulated –detected by MS 93.80% 92.20% [109]Case-control Serum Amino acids (panel) 8 amino acids –detected by LCMS/MS 65.00% 95.00% [110]Case-control Serum Amino acids, fatty acids, carbohydrates 13 metabolites –detected by LCMS/MS 96.00% 80.00% [111]Case-control Serum Metabolite panel 2-hydroxy-butyrate,aspartic acid,kynurenine,cystamine – detected by GC-MS 83.10% 81.00% [112]Case-control Serum Lipid metabolites(panel)Palmitic amide,oleamide,hexadecaneodioic acid, octadecanoic acid, eicosatrienoic acid, LPC(18:2),LPC(20:4),LPC(22:6), myristic acid, LPC(16:0) –detected by ion cyclotron resonance MS 98.10% 100.00% [113]Case-control Serum Panel of hydroxylated polyunsaturated ultra long-chain fatty acids C28H46O4,C28H48O4 and C28H50O4, all downregulated –detected by LCMS/MS and nuclear MR 75.00% 90.00% [114]Case-control Serum Multiple metabolites(panel)11,14-eicosadienoic acid, 12a-hydroxy-3-oxocholadienic acid,12-ketodeoxycholic acid, 12-ketotetrahydroleukotriene Β4, 13-cis-retinoic acid, 1bhydrocholic acid, 1-methylhistamine, 1-monopalmitin, 2,3-dihydroxybutanoic acid, 24-hydroxycalcitriol –detected by GCTOFMS and UPLCQTOFMS 83.70% 91.70% [115]
99.30% 93.80% [116]Case-control Plasma Amino acids, fatty acids, carbohydrates 8 metabolites –detected by CTTQMS Case-control Plasma Choline-containing phospholipids(panel)Total saturated lysophosphatidylcholines (LPCs), 18:2 LPC and sphingosylphosphor ylcholine – detected by LC-MS/MS 88.30% 80.00% [117]Case-control Plasma Choline-containing phospholipids(panel)Total saturated lysophosphatidylcholines (LPCs) and the difference between 18:2 LPC and 18:1 LPC –detected by LC-MS 82.00% 93.00% [118]Case-control Dried blood Amino acids and acylcarnitines(panel)C16, Arg, C4/C8,C5/C3, Val,Phe/Tyr, Ala,C4/C3 – detected by direct infusion MS 81.20% 83.90% [119]Case-control Urine Polyamines N1, N12-diacetylspermine –detected by ELISA 75.80% 96.00% [120]Case-control Urine Polyamines and amino acid metabolites N1, N12-diacetylspermine and kynurenine –detected by LC-MS 80.00% 80.00% [121]Case-control Urine Amino acids and acetoacetate (panel)Alanine, glutamine,aspartic acid and acetoacetate –detected by PMRS 87.50% 91.30% [122]Case-control Urine Nucleosides (panel) 5-hydroxymethyluraci l and 8-oxo-7,8-dihydroguanine –detected by UPLCMS/MS 78.60% 75.00% [123]Case-control Urine Nucleosides (panel) Cytidine, 3-methylcitidine, 1-methyladenosine, 2-deoxyguanosine,adenosine, inosine –detected by HPLCMS/MS 69.00% 98.00% [124]Case-control Urine Metabolite panel Citrate, Hippurate,p-cresol, 2-aminobutyrate,myristate, putrescine and kynurenate -detected by UPLCQTOFMS 97.50% 100% [125]Case-control Urine Nucleosides (panel) Adenosine, N4-acetylcytidine,cytidine, guanosine,inosine, 1-methyladenosine, 1-methylguanosine, 1-methylinosine, 2-methylguanosine,2,2-methylguanosine,N6-methyladenosine,uridine, 3-methyluridine+5-methyluridine,pseudouridine –detected by reverse phase HPLC 76.90% 90.40% [126]
Case-control Urine Nucleosides (panel) Adenosine, N4-acetylcytidine,cytidine, guanosine,inosine, 1-methyladenosine, 1-methylguanosine, 1-methylinosine, 2-methylguanosine,2,2-methylguanosine,N6-methyladenosine, 5-methyluridine,pseudouridine,uridine – detected by column switching HPLC 71.00% 96.00% [127]
One remarkable common feature of all the studies listed in Table 5 is the obligatory presence ofFusobacterium nucleatum(F. nucleatum) as one of the components of all tested panels. Indeed,F. nucleatum, an anaerobic oral commensal, is now identified as a pathogenetic factor contributing to multiple disorders comprising among others inflammatory bowel disease and CRC[19,148,149]. This interesting diagnostic approach is being actively investigated; however, further studies are necessary to firmly establish the value of the gut microbiome in non-invasive CRC detection.
NON-INVASIVE BIOMARKER TESTING USE IN CRC SCREENING TODAY AND FUTURE CHALLENGES
The existing plethora of potential non-invasive approaches to CRC detection briefly reviewed in this paper looks impressive in terms of numbers, but often disappointing in terms of outcome. Most of the published results clearly fail to transform into diagnostic or screening tests that would be highly sensitive and specific, simple to perform and not associated with excessive cost. As a matter of fact, the choice of available biomarker-based tests practically used for CRC screening remains strictly limited. Today FIT is by far the most popular option[2,9,31]owing to its relative simplicity and affordability. The recently introduced and widely advertised multitarget Cologuard?stool test or Epi proColon test targetingSEPT9methylation in plasma, albeit approved for clinical use, are technically complex and prohibitively expensive. Comparative studies addressing the health economics of CRC screening have demonstrated that the multitarget stool test, being more cost-effective that no screening, is significantly less cost-effective when compared to the FIT or invasive endoscopic testing[150-152]. Likewise, methylated SEPT9 detection in plasma samples[153]is clearly less cost-effective than the FIT. Considering a unit cost of $8 for the FIT(sampling kit and analysis only), Lansdorp-Vogelaaret al[154]concluded that a biomarker-based test that detects CRC with higher levels of sensitivity and specificity(up to 100%) should never be more expensive than $57 to be cost-effective. These estimates seem to indicate that in practical terms the FIT is currently the most costeffective test for non-invasive CRC screening. Other authors argue that a highly specific non-invasive biomarker with an improved sensitivity for advanced adenomas(that progress to CRC) would probably be cost-effective at higher threshold costs[155],but the $600 price tag currently attached to Cologuard?is obviously excessive.
In any case, it is apparent that the FIT is not a perfect screening test. Its specificity reaching 95% is sufficiently high to be deemed satisfactory, but the sensitivity of this test remains relatively modest[31]. There is, however, an opinion that repeated FIT testing with one-year intervals may compensate for the lack of sensitivity[12].Moreover, accurate identification of individuals with different levels of CRC risk could lead to creating objective approaches to risk stratification and personalised screening[12,155,156].
The effectiveness of a screening strategy is defined not only by screening test performance characteristics, but also by screening participant adherence[12]. One additional practical problem in CRC screening programmes employing faecal tests is insufficient screening uptake[157,158]that often results from participants’ reluctance to collect stool samples[159,160]. The use of non-invasively collected colorectal mucus samples[24,138]in FIT-like tests can help solve this problem, but this new approach remains to be thoroughly evaluated, and this will require large comparative randomised trials that usually take several years to complete[155]. The existingcombination of the FIT and confirmatory colonoscopy is the strategy of choice today,and its further optimisation is currently regarded as the main factor in improving CRC screening effectiveness.

Table 5 Non-invasive faecal bacterial biomarkers used for colorectal cancer detection
The present strong position of the FIT as the test of choice for non-invasive CRC screening will certainly be temporary as this test has one intrinsic deficiency that is impossible to eliminate. The FIT detects blood, which is shed but not produced by tumours, and bleeding may not occur in some CRC patients. For this reason, FIT sensitivity will never approach 100%, and it is likely that this target will become achievable only when a screening test employing CRC-specific biomarker(s) is developed. As no single biomarker detectable in all colorectal tumours has been identified so far, multitarget strategies combining either multiple markers of the same type or different assays (such as Cologuard?) emerge as CRC screening options advocated by some experts. However, these complex assays usually require sophisticated laboratory equipment and are laborious and expensive. Although future technological advances can help in eliminating these deficiencies, the search for more reliable and easily detectable single CRC biomarkers should continue.
It can be expected that rapid progress in cancer biomarker research accompanied by accelerated development of new non-invasive tests promises forthcoming breakthroughs in CRC screening and prevention of this disease.
 World Journal of Gastrointestinal Oncology2020年2期
World Journal of Gastrointestinal Oncology2020年2期
- World Journal of Gastrointestinal Oncology的其它文章
- Caffeine and its main targets of colorectal cancer
- FOLFIRINOX vs gemcitabine/nab-paclitaxel for treatment of metastatic pancreatic cancer: Single-center cohort study
- Prognostic scoring system for synchronous brain metastasis at diagnosis of colorectal cancer: A populationbased study
- Neuropathy experienced by colorectal cancer patients receiving oxaliplatin: A qualitative study to validate the Functional Assessment of Cancer Therapy/Gynecologic Oncology Group-Neurotoxicity scale
- Clinical value evaluation of serum markers for early diagnosis of colorectal cancer
- Yttrium-90 radioembolization for unresectable hepatic metastases of breast cancer: A systematic review
