Expression Profiles and Protein Complexes of Starch Biosynthetic Enzymes from White-Core and Waxy Mutants Induced from High Amylose Indica Rice
CHEN Yaling , PANG Yuehan BAO Jinsong
(1Institute of Nuclear Agricultural Sciences, College of Agriculture and Biotechnology, Huajiachi Campus, Zhejiang University,Hangzhou 310029, China; 2College of Life Sciences, Jiangxi Normal University, Nanchang 330022, China)
Abstract: Physicochemical properties of endosperm starches in milled rice determine its cooking and eating quality. Amylose is synthesized by granule-bound starch synthase I (GBSSI), whilst amylopectin is synthesized by the synergistic activities of starch synthases (SSs), branching enzymes (BEs) and debranching enzymes (DBEs). However, the complexes formed by starch biosynthetic enzymes are not well characterized. Gene expression profiles and protein complexes were determined in white-core(GM645) and waxy (GM077) mutants derived from a high amylose indica rice Guangluai 4 (GLA4). In GM645, genes including AGPS1, GBSSI, SSIIa, BEI, BEIIa, BEIIb, PUL, ISA1 and SP were significantly downregulated during seed development. In GM077, the expression levels of AGPL2, AGPS1, AGPS2b,SSIIIa, BEI, PUL and ISA1 were significantly upregulated. Co-immunoprecipitation assays revealed interactions of SSs-BEs, SSs-PUL and BEs-PUL in developing seeds. However, weak SSI-SSIIa interaction was detected in GM077, whilst SSI-PUL interaction was absent. Weak interaction signals for SSI-SSIIa, SSIIa-BEI, SSIIa-BEIIb, BEI-BEIIb and SSI-BEI were also observed in GM645. These results suggest that the protein-protein interactions for starch biosynthesis are modified in mutants, which provides insight into the mechanisms of starch biosynthesis, particularly in indica rice.
Key words: amylose; endosperm mutant; indica rice; protein-protein interaction; starch biosynthetic enzyme; waxy rice
The physicochemical properties of endosperm starch in milled rice determine its cooking and eating quality(Vandeputte and Delcour, 2004; Bao, 2012, 2019; Xu et al, 2018). Starch is the major storage material in the rice endosperm, accounting for 80%-90% of the seed weight, and is composed of amylose and amylopectin(Vandeputte and Delcour, 2004; Du et al, 2019). Starch mutants are defective in either amylose or amylopectin biosynthesis and have been used for the molecular characterization of key enzymes and regulatory factors during starch synthesis. Defects in granule-bound starch synthase I (GBSSI) produce a waxy (wx)endosperm composed of amylose-free starch grains,confirming the essential role of GBSSI in amylose synthesis (Jeng et al, 2009; Zhang et al, 2012). The functions of other enzymes in amylopectin synthesis have also been shown in starch mutants such as ADP-glucose pyrophosphorylases (AGPases), soluble starch synthases (SSs), branching enzymes (BEs), and debranching enzymes (DBEs) (Nishi et al, 2001;Fujita et al, 2006, 2007, 2009; Nakamura et al, 2010;Toyosawa et al, 2016). Furthermore, mutations in these genes exhibit abnormal features of starch storage in the endosperm. For example, SSIIIa mutants produce a chalky interior, and the amylose content and physicochemical properties of the starch granules(SGs) are influenced (Fujita et al, 2007; Ryoo et al,2007; Bao, 2019). Loss-of-function mutations in BEIIb lead to a white-core endosperm, with altered amylopectin structures and gelatinization properties of the SGs (Nishi et al, 2001; Sawada et al, 2018).Isoamylase1 (ISA1)-deficient mutants (isa1) are also termed as sugary mutants in rice (sug-1) and maize(su1) (James et al, 1995; Kubo et al, 2005). Residual SGs in the isa1 mutants are of abnormal size, shape and number (Kubo et al, 2010; Utsumi et al, 2011). Studies in starch mutants have enhanced our understanding of the mechanisms for starch biosynthesis.
Recent works also indicate that the protein complexes of starch synthesis isozymes may exist during seed development in wheat, maize, barley and rice(Hennen-Bierwagen et al, 2008; Tetlow et al, 2008;Ahmed et al, 2015; Crofts et al, 2015, 2017), but complexes in the mutant endosperm may differ from those in wild type (Liu et al, 2009, 2012a; Crofts et al,2018). Tetlow et al (2004) first reported a multi-enzyme complex of BEIIb, BEI and starch phosphorylase (SP)in the amyloplasts of developing wheat endosperm.Subsequently, other enzyme complexes, including SSI,SSIIa, SSIIIa, BEIIa and/or BEIIb, in various combinations have been identified (Hennen-Bierwagen et al, 2008; Ahmed et al, 2015). In developing maize seeds, Liu et al (2009) reported that a major protein complex consists of SSI, SSIIa and BEIIb, but the null amylose extender mutant of SBEIIb contains a novel protein complex comprised of SSI, SSIIa, SBEI,SBEIIa and SP. Liu et al (2012b) further reported that a point mutation in SSIIa in a su2-mutant of maize causes the loss of SSI and SBEIIb, in addition to SSIIa from SGs. In the SSIIa mutants (sex6) of barely, no protein complexes involving SBEIIa or SBEIIb are detected in amyloplasts (Ahmed et al, 2015). The BEII isoform in the formation of novel protein complexes is substituted by BEI and SP (Ahmed et al, 2015). These observations imply that alterations in the granule proteome arise from genetic mutations or the downregulation of specific genes gives rise to variations in protein complexes in the amyloplasts.
Rice has 10 SS isoforms, 3 BE isoforms, and 4 DBE isoforms. These isozymes have been characterized in studies of starch mutants, predominantly from japonica varieties, including Nipponbare, Taichung 65 and Kinmaze (Sawada et al, 2018). Co-immunoprecipitation(Co-IP) assays of SSs, BEs and DBEs provide direct evidence of the formation of multi-enzyme complexes in soluble extracts of Nipponbare endosperm (Crofts et al, 2015; Chen and Bao, 2016). However, compared to japonica varieties, indica rice varieties have active SSIIa isozymes (Nakamura et al, 2005) and high expression level of GBSSI (Bligh et al, 1998; Cai et al,1998; Dobo et al, 2010). Challenges remain regarding the mechanisms for starch synthesis in indica rice,including whether the elevated expression of GBSSI affects the activities of other enzymes (Bao, 2012).Whether novel protein complexes are present in indica rice due to the active SSIIa isozyme is also unknown.Whether the activities of enzymes and protein complex change during starch synthesis in indica mutants is also uncharacterized.
In previous studies, two stable starch mutants were isolated from high amylose indica rice, Guangluai 4(GLA4) (Kong et al, 2014). One is a waxy mutant(GM077) with an opaque endosperm and very low apparent amylose content (2.6%). The other is a whitecore mutant (GM645) with a chalky endosperm in the center of the grain. The physicochemical properties of the starch and chain length distributions of the amylopectin extracted from the two mutants differ(Kong et al, 2014). In this study, the expression levels of genes and enzymes involved in starch synthesis,including AGPases, GBSSI, SSs, BEs and DBEs, in developing rice endosperms were measured by qRT-PCR and western blotting. Co-IPs were performed to characterize the protein complexes formed in the mutants. Our results provided direct evidence for changes in protein complexes in rice mutants and enhanced our understanding of starch biosynthesis in indica rice.
MATERIALS AND METHODS
Materials
High amylose indica rice GLA4 and two stable endosperm mutants (GM077 and GM645) were grown in the field at the Zhejiang University farm, Hangzhou,China. Individual panicles were labeled during flowering. Developing seeds at 5, 10 and 15 d after flowering (DAF) were collected, and immediately frozen on ice and stored at -80 °C. Antisera against rice GBSSI, SSI, SSIIa, BEI, BEIIb and PUL were a kind gift of Dr. Naoko FUJITA at Akita Prefectural University, Akita, Japan.
RNA extraction and quantitative real-time PCR(qRT-PCR)
Total RNAs from the rice endosperm at different developmental time points were isolated according to the manufacturer’s protocols using the SV Total RNA Isolation System (Promega, Beijing, China). cDNA was synthesized using the First-Strand Synthesis cDNA kit (Promega, Beijing, China). Gene-specific primers of starch synthesis related genes were used in qRT-PCRs,referring to Ohdan et al (2005). Additionally, the housekeeping gene Actin was used as an internal control to normalize cDNA levels in each sample.qRT-PCRs were performed using StepOneTM&StepOne PlusTMReal-Time PCR Systems (ABI, USA)accompanying with SYBR?Premix Ex TaqTM(TaKaRa,Dalian, China). Relative gene expression was calculated using the 2-ΔΔCTmethod (Jain et al, 2006). Measurements at each time point were performed in triplicate.
Extraction of soluble proteins and starch granulebound proteins
The isolation of soluble proteins was performed as previously described (Chen and Bao, 2016). Proteins bound with starch granules were extracted according to Fujita et al (2006), with minor modifications. Following protein extraction, the starch was washed twice with three volumes of cold sodium dodecyl sulfate (SDS)solution containing 55 mmol/L of Tris-HCl (pH = 6.8),2.3% of SDS, 5% of 2-mercaptoethanol and 10% of glycerol, to remove residual proteins attached to the surface (Fujita et al, 2006). Residual pellets (starch granules) were washed with 1 mL of distilled water and 1 mL of acetone twice. Extracts were dried using a freeze drier. Equivalent amounts of starch (50 mg)were boiled in 10 volumes of SDS solution for 10 min.After cooling, 20 volumes of SDS solution were added with stirring, and samples were centrifuged at 12 000 ×g for 10 min at 4 °C. Supernatants were assessed for starch granule-bound proteins. Protein concentrations were measured on a NanoDrop 2000 spectrophotometer(Thermo, Canada).
Co-immunoprecipitation (Co-IP) assay
Co-IP assays were performed as described by Crofts et al(2015) and Chen and Bao (2016) with some modifications. Soluble proteins (200 μL, 10 mg/mL)were mixed with different volumes of primary antibodies (15 μL anti-SSI, 20 μL anti-SSIIa, 10 μL anti-BEI, 15 μL anti-BEIIb, or 15 μL of anti-PUL) for 2 h at 4 °C. A 200 μL aliquot of reconstituted 50%protein A-sepharose resin (TransGen, China) was added and incubated on a rotator for 3 h at 4 °C.Protein A-sepharose-antibody-protein complexes were centrifuged at 6 000 × g for 3 min at 4 °C and supernatants were discarded. The resins were washed eight times with PBS (137 mmol/L NaCl, 10 mmol/L Na2HPO4, 2.7 mmol/L KCl, 1.8 mmol/L KH2PO4, pH 7.4). Bound proteins were released by boiling for 10 min in 1× SDS buffer. After centrifugation at 12 000 r/min for 3 min, 10 μL of the lysates were used for western blotting.
Western blotting
Proteins were resolved by 10% SDS-PAGE (SDSpolyacrylamide gel electrophoresis), and transferred onto polyvinylidene fluoride (PVDF) membranes using a transblotter. Western blotting procedure was performed as described by Crofts et al (2015). Blots were repeated at least three times, while blots for the Co-IPs were performed at least two times.
Data analysis
Western blots were quantitated using the Image J software. t-tests of the differences in expression levels between the mutant and wild type were performed using the SPSS 20.0 software (SPSS, Inc., Chicago IL,USA).
RESULTS
Expression profiles of starch synthesis genes
Previous studies showed that major starch genes including AGPL1, AGPL2, AGPS1, AGPS2b, GBSSI,SSI, SSIIa, SSIIIa, BEI, BEIIa, BEIIb, ISA1, PUL and SP are expressed in rice endosperms (Duan and Sun,2005). The expression levels of these genes in two endosperm mutants (GM077 and GM645) during the developing of rice grains at 5, 10 and 15 DAF were examined by qRT-PCR.
Compared to the wild type GLA4, the 14 genes displayed four different expression profiles in GM077.The expression of GBSSI was downregulated, whereas the expression levels of AGPL2, AGPS1, AGPS2b, SSIIIa,BEI, PUL and ISA1 were significantly upregulated(Fig. 1). The expression levels of SSI and BEIIb were downregulated at 5 DAF but subsequently upregulated.The expression levels of AGPL1, SSIIa, BEIIa and SP were upregulated from 5 to 10 DAF but subsequently downregulated. These data revealed that the expression levels of most starch synthesis genes were upregulated in GM077 seeds during the grain-filling stage.
In the white-core endosperm mutant GM645, the expression levels of AGPS1, GBSSI, SSIIa, BEI, BEIIa,BEIIb, PUL, ISA1 and SP were significantly downregulated during endosperm development. The expression levels of AGPS2b, SSI and SSIIIa were downregulated from 5 to 10 DAF but subsequently upregulated (Fig. 1).
Accumulation of starch synthesis related proteins
To assess the levels of protein accumulation in the two endosperm mutants (GM077 and GM645), we performed western blotting of the wild type and the two endosperm mutants during rice endosperm development with various antibodies. As shown in Fig. 2-A, GBSSI protein was undetectable in rice endosperm of GM077 during grain filling. SSIIa protein at 5 DAF increased in GM077, which was comparable to GLA4 at 10 and 15 DAF. There were no significant changes for SSI,BEI and BEIIb from 5 to 15 DAF (Fig. 2-B).
In the white-core endosperm mutant GM645, a significant reduction in SSIIa, BEI, BEIIb and PUL expression was observed. SSI at 10 and 15 DAF in GM645 was similar to that in GLA4 (Fig. 2-B). These results suggested that the white-core mutation reduced the levels of soluble proteins in relation to starch biosynthesis in the endosperms.
Analysis of protein-protein interactions
To investigate possible interacting partners amongst starch biosynthetic isozymes in the wild type (GLA4)and mutants of indica rice, Co-IPs were performed using soluble endosperm extracts at 10 DAF (Table 1 and Fig. 3).
Strong pairwise associations through Co-IPs were observed for SSI-SSIIa, SSI-BEIIb, SSI-PUL, SSIIa-BEI, SSIIa-BEIIb, BEI-BEIIb, BEI-PUL and BEIIb-PUL in the GLA4 endosperm. Interactions were observed for SSI-BEI and SSIIa-PUL (the first acronym:antibodies used for immunoprecipitation; the second acronym: antibodies detected by western blotting),whilst the reciprocal Co-IPs were relatively weaker.
In the GM077 endosperm, strong pairwise associations were obtained by reciprocal Co-IPs for SSI-BEI,SSI-BEIIb, SSIIa-BEI, SSIIa-BEIIb, SSIIa-PUL, BEIBEIIb, BEI-PUL and BEIIb-PUL. Compared to GLA4,the pairwise interaction signals of SSI-SSIIa were weaker. Of note, SSI-PUL interactions were not detected in the GM077 endosperm.
In the GM645 endosperm, the interactions of SSIBEIIb, SSI-PUL, BEI-PUL, BEIIb-PUL and SSIIa-PUL were similar to the GLA4 endosperm. Pairwise interactions for SSI-SSIIa, SSIIa-BEI, SSIIa-BEIIb and BEI-BEIIb in the GM645 endosperm were weaker than those observed in the wild type. Furthermore, a clear, but less intense signal for SSI-BEI was obtained from only one side of Co-IP in the GM645 endosperm.
DISCUSSION
Protein complexes of starch biosynthetic enzymes from high amylose indica rice GLA4
Interactions between starch biosynthetic isozymes have been shown in wheat, maize, barley and japonica rice developing endosperms (Hennen-Bierwagen et al,2008; Tetlow et al, 2008; Ahmed et al, 2015; Crofts et al,2015). Co-IP assays revealed associations of SSs-BEs,BEIIa-Pho1, and pullulanase-type DBE-BEI present in japonica rice Nipponbare (Crofts et al, 2015; Hayashi et al, 2018; Miura et al, 2018), which possesses inactive SSIIa and lower GBSSI expression levels compared to indica rice. In high amylose indica rice GLA4 that possesses active SSIIa and higher GBSSI expression,SSI-BEs, SSI-PUL and PUL-BEI complexes were similar to japonica rice Nipponbare (Crofts et al, 2015;Hayashi et al, 2018), but SSI-SSIIa, SSIIa-BEs and PUL-SSIIa were found only in GLA4 (Fig. 3 and Table 1). These discoveries provide a basis for the comprehensive analysis of protein complexes of starchbiosynthetic enzymes in rice, although the possibility remains that the differences arise not only from each plant-specific function, but from differences in the experimental approaches, such as the choice of endosperm development and whether amyloplasts or whole-cell extracts are used as the starting material.
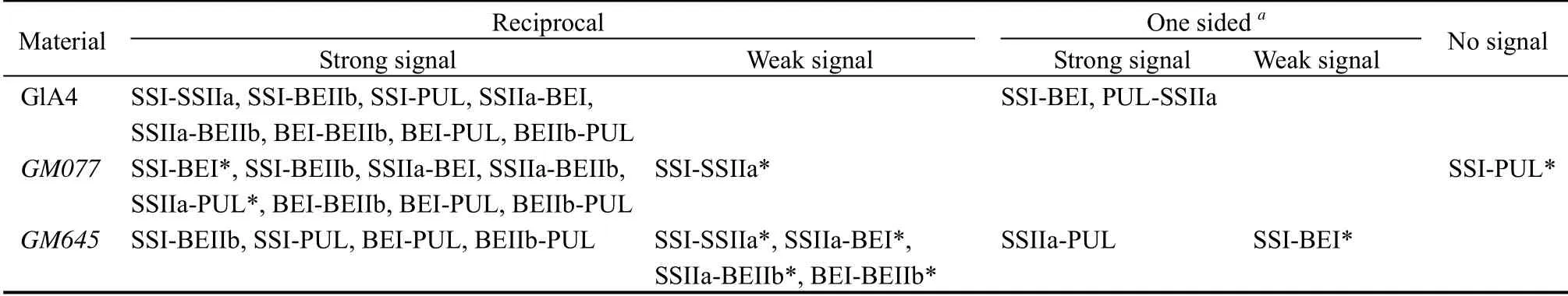
Table 1. Comparison of protein-protein interactions among starch synthetic related enzymes in wild type and mutant endosperms determined by co-immunoprecipitation assay.
Protein complexes of starch biosynthetic enzymes from waxy mutant GM077
The waxy mutants of cereals and the responsible gene(GBSSI) were discovered many years ago (Bligh et al,1998; Cai et al, 1998; Dobo et al, 2010). Due to the loss of amylose accumulation, reallocation of the carbon source to amylopectin synthesis in GM077 has been reported by Zhang et al (2012), who found that the elevated expression levels of AGPS and ISA1 in the rice waxy mutants drive carbon flux from amylose to amylopectin synthesis. In this study, qRT-PCR analysis indicated that GM077 was a waxy mutant with high expression levels of 11 amylopectin synthetic related genes, and that higher AGPS1 and ISA1 expression levels were detected during the whole endosperm developmental stage compared to the wild type GLA4,which is consistent with Zhang et al (2012). The mechanism behind the elevated expression of AGPS and ISA1 in the rice mutants remains unclear. The α-1,4-glucosidic link chains of both amylose and amylopectin are elongated through the addition of the glucose moiety from ADP-glucose. One possibility is that the extra AGPase is required in GM077 to convert Glc1-P to ADP-glucose in the cytosol. ISA1 is particularly important for amylopectin formation, and therefore, it is unsurprising that ISA1 expression in GM077 significantly increased when excess amylopectin was produced in the endosperm (Utsumi et al, 2011;Sun et al, 2015).
Novel complexes of starch biosynthetic enzymes have been reported in rice mutants lacking SSI and BEIIb (Crofts et al, 2018), but reports on starch biosynthetic protein complexes in rice waxy mutants are sparse. Co-IP analysis indicated the pairwise interaction signals of SSI-SSIIa in the waxy mutant GM077 were weaker compared to GLA4. The weak interaction may be due to the low levels of SSIIa in GM077 compared to GLA4, although the expression level and protein content of SSI were higher in GM077 than in GLA4 (Fig. 2). Moreover, it was interesting to note that no SSI-PUL interaction was detected in reciprocal Co-IPs in GM077 endosperm.Western blotting also detected a low amount of PUL protein (Fig. 2-B). However, the expression of ISA1 in the GM077 endosperm was significantly higher than that of GLA4 (Fig. 1). Fujita et al (2009) provided evidence that the role of PUL is to supplement ISA when it is absent. Utsumi et al (2011) reported that the ISA1-ISA1 homomer is the only functional form in rice endosperm. Although interaction of SSI and ISA has not been reported, it is worth noting that a close correlation between ISA and SS activities during starch accumulation occurs in Arabidopsis leaves,suggesting functional or physical interaction exists between these two enzyme classes (Pfister et al, 2014).This may explain the loss of SSI-PUL in GM077 due to increased function of the ISA1-ISA1 homomer or the interaction between SSI and ISA1.
Protein complexes of starch biosynthetic enzymes from white-core mutant GM645
In the white-core mutant GM645, reductions in expression level of starch synthesis genes, including
AGPS1, GBSSI, SSIIa, BEI, BEIIa, BEIIb, PUL, ISA1 and SP, were observed (Fig. 1). Western blot analysis revealed an extreme reduction of BEI, BEIIb and PUL(Fig. 2). Co-IPs indicated that pairwise interactions between BEI-SSIIa and BEI-BEIIb were weaker than those of wild type. Furthermore, weaker signal of SSI-BEI was obtained from only one of the Co-IPs in the GM645 endosperm.
Gene mutations have not been characterized in GM645. Previous studies isolated and functionally characterized four white-core endosperm mutants,including flo4, flo5, rsr1 and flo12 (Kang et al, 2005;Ryoo et al, 2007; Fu and Xue, 2010; Zhong et al,2019). Amongst them, flo12 shows the reduced expression of starch synthesis genes, including SSI,SSIIb, SSIII, SSIV, AGPL, ISA1 and BEI (Zhong et al,2019). The expression levels of SSI and SSIIIa in GM645 remained unchanged, so GM645 is not mutated at the flo12 locus. The floury-white endosperm flo2 exhibits reduced levels of BEI in the developing rice endosperm, along with decreased levels of other starch-synthetic enzymes, including AGPase, GBSS,SS and BEIIb (Kawasaki et al, 1996). She et al (2010)reported that the flo2 mutant shows lower expression of genes involved in the production of storage starch in the endosperm. Wu et al (2015) reported that three flo2 mutants perturb the expression of starch synthesisrelated genes including OsAGPL2, OsAGPS2b, OsGBSSI,OsBEI, OsBEIIb, OsISA1 and OsPUL. Consistent with these findings, both gene expression and western blot analysis indicated that starch synthesis genes are down-regulated in GM645 (Figs. 1 and 2). According to the mutant phenotypes, GM645 had higher numbers of short and long chains consisting of 6-9 degrees of polymerization (DP) and ≥ 44 DP, respectively (Kong et al, 2014), but flo2 has lower numbers of short and long chains (She et al, 2010). Mutations in GM645 await further map-based cloning, but the mechanisms by which these protein complexes influence starch biosynthetic enzymes now require further investigation.
Proposed models for starch synthesis in high amylose indica rice, waxy mutants and white-core mutants
Functional redundancy has been observed for mutants lacking specific rice starch biosynthetic enzymes(Nakamura, 2002; Fujita, 2014). GBSSI is primarily involved in amylose biosynthesis. SSI elongates short chains (8-12 DP) of amylopectin generated by BEIIb,and the 8 DP chains can be elongated to form intermediate intra-cluster chains (12-24 DP) by SSIIa.SSIIIa generates long chains (> 30 DP) connecting multiple clusters of amylopectin. ISA and PUL directly debranch improper branches. In our previous studies, waxy mutant GM077 had more chains with 6-10, 18-33 and > 44 DP in addition to higher gelatinization temperature, whilst white-core mutants GM645 had more chains with 6-9, 22-35 and > 44 DP as well as with lower gelatinization temperature than GLA4 (Kong et al, 2014). Through the analysis of the chain length distribution of amylopectin, expression profiles and protein complexes from GLA4, GM077 and GM645, mechanisms for the formation of enzyme complexes involved in starch biosynthesis in the endosperms of high amylose indica rice, waxy mutant and white-core mutant were proposed (Fig. 4).
In the GLA4 endosperm that possesses higher levels of SSIIa expression, amylose and amylopectin are elongated by the addition of the glucose moiety from ADP-glucose (Fig. 4-A). The trimeric complex of BEI, SSI and SSIIa or BEIIb, SSI and SSIIa binds to the glucan, branches and elongates the polymer,generating intermediate intra-cluster chains (12-24 DP) and acting as a ‘glucan-chaperone’ (Tetlow et al,2015). SSI-SSIIa contributes to the initial short chain elongation of 6-7 DP produced by BEIIb to form 12-24 DP chains. SSIIIa activity, which is abundant at the starch filling stage, elongates the intermediate chains to 30 DP. The complex may be unable to proceed as a result of the formation of disorganized branches that lead to steric hindrance. ISA1-ISA1 homomers or the complexes of BEs-SSs-PUL remove the disorganized branches (Fig. 4).
In the GM077 endosperm, higher SSIIIa expression may increase the synthesis of intermediate chains that lead to BEI-SSI-SSIIa from 18-33 DP chains (Fig. 4).BEIIb-SSI or SSI branches and elongates the polymer,generating intermediate chains (8-12 DP) due to the weaker interaction of SSI-SSIIa (Table 1). Disorganized branches were removed by ISA1-ISA1 homomers due to the lack of SSI-PUL complex in GM077 (Table 1).
In the GM645 endosperm, complexes of BEI-SSIIa,BEI-BEIIb and SSI-BEI were weaker, but SSI-BEIIb interactions were comparable to GLA4. The complex of SSI-BEIIb forms more chains with 6-9 DP (Fig. 4).Similar SSIIIa expression may elongate the short chains to 22-35 DP. Disorganized branches were removed by the same complexes as wild type.
It is noteworthy that Pang et al (2018) identified phosphorylated proteins (SSIIa, SSIIIa, BEI, BEIIb,PUL and SP) related to starch synthesis in indica rice.We hypothesized that the formation of multiple enzyme complexes are related to the phosphorylation of rice starch biosynthesis related enzymes. As in wheat and maize, complexes are catalyzed as a result of phosphorylation (Tetlow et al, 2004, 2008; Liu et al,2009). Future studies focusing on the cloning and functional characterization of genes for the white-core mutant, validation of the mechanisms for the differences in protein-protein interactions, and assessments of the involvement of protein phosphorylation during protein complex formation in amylopectin synthesis between the wild type and mutant endosperms are required.
ACKNOWLEDGEMENTS
This work was financially supported by the National Key Research and Development Program of China(Grant No. 2016YFD0400104), and the Natural Science Foundation of China (Grant Nos. 31800640 and 31871531).
- Rice Science的其它文章
- INSTRUCTION FOR CONTRIBUTORS
- Rhizosphere Aeration Improves Nitrogen Transformation in Soil,and Nitrogen Absorption and Accumulation in Rice Plants
- Rice Heavy Metal P-type ATPase OsHMA6 Is Likely a Copper Efflux Protein
- OsSRK1, an Atypical S-Receptor-Like Kinase Positively Regulates Leaf Width and Salt Tolerance in Rice
- Genetic Relationship and Structure Analysis of Root Growth Angle for Improvement of Drought Avoidance in Early and Mid-Early Maturing Rice Genotypes
- Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast

