Pharmacologic mechanisms mining and prediction of Xiaoer Qixing Cha Formulae in the treatment of infantile functional dyspepsia based on chemical analysis by UPLC-QTOF/MS and interactive network pharmacology
Mei-Qi Wang,Zeren Dawa,Yu-Feng Yao,Fang-Le Liu,Run-Jing Zhang,Zi-Yuan Wang,Chen-Chen Zhu*Institute of Clinical Pharmacology, Guangzhou University of Chinese Medicine, Guangzhou, China. Tibetan Traditional Medical College,Lasa,China. The First Affiliated Hospital of Chinese Medicine, Guangzhou University of Chinese Medicine,Guangzhou,China.
Background
Infantile function dyspepsia (IFD), one of the most common pathological states in pediatrics, is a gastrointestinal function disorder with upper abdominal pain and distension, early fullness, nausea, vomiting and burning sensation of superior belly 0. The above related symptoms seriously affect the children's food intake, and lead to long-term inadequate nutrition intake, high incidence of malnutrition, growth and development retardation, and even neurosis, anxiety disorders and other mental disorder in children. Although its pathogenesis still keeps unknown, several ways, such as gastrointestinal motility disorder,visceral hypersensitivity,abnormal gastric acid secretion, Hp infection and psychosocial factors have been considered to be highly related to IFD 1. Clinically, effective treatments for IFD mainly refer to promotion of gastrointestinal motility,inhibition of gastric acid, improvement of intestinal mucosal immune function and modulation of regulatory function of central nervous system (CNS), brain-gut axis and enteric nerve (ENS) 3.Diverse types of synthetic drugs have been used to relieve IFD in clinic.However,a single type of drug with limitation can hardly satisfy the treatment demand for its complicated pathogenesis 4.For the complexity monogenesis of IFD and high tendency of recurrence, exploring new therapeutic methods are urgently needed and under intensive investigation.
As a complementary therapy, Chinese medicinal formulae have been applied with apparent safety and efficacy for thousands of years. According to the unique compatibility theory of traditional Chinese medicine,herbal drugs are jointly used on the basis of their flavor and meridian tropism with appropriate amount which has been proven the curative effect and reduce toxicity of the Formulae56.With the superiority and individuality of multiple ingredients, multiple targets and synergistic effects, Chinese medicinal Formulae has multiple functions of synchronously treated complicated diseases 7.
Xiaoer Qixing Cha Formulae (XQCF) is a well-known pediatric prescription in China and has been widely applied clinically to relieve dyspepsia, improve appetite,clear away heat and toxic materials from body, calm and soothe the nerves, and eliminate anxiety originated from IFD 8 . XQCF is consisted of seven herbs, namely Yiyiren (Coix lacryma-jobi L. var. mayuen (Roman.)Stapf), Daoya (Oryza germinatus L.), Shanzha(Crataegus pinnatifida Bge.), Danzhuye (Lophatherum gracile Brongn),Gouteng(Uncaria rhynchophylla(Miq.)Miq. ex Havil.), Chantui (Cicada slough) and Gancao(Glycyrrhiza uralensis) 9. Modern pharmacological studies have revealed that XQCF possess prominent laxative activity and effectively protects the colonic mucosal barrier 10. However, the pharmacological mechanisms on IFD of XQCF remain unknown.
The complexity components and their group effects in Chinese medicinal Formulae result in some problems such as unclear mechanisms and unclear substance bases.There for a systematic analysis of its chemical constituents and targets will help to clarify the pharmacological mechanisms of traditional Chinese medicine (TCM) Formulae. UPLC-QTOF/MS is an effective method for rapid separation and identification of chemical components in complex system of TCM 11-12].Interactive Network Pharmacology is a systemic and comprehensive method of exploring and mining to illuminate scientific bases and potential mechanisms of TCM 13-14. On the bases of the multiple network of herbs-targets-disease-protein, it realizes prediction of drug targets and improvement of drug discovery efficiency from overall aspects. It has been increasingly applied to exploring and predicting the pharmacological mechanisms of TCM. However, the data sources of most previous studies were mainly from chemical components reported in the literature 15and the requirements of extraction processes of herbs are different in Formulaes.That leads to the inconformity of the chemical composition of the prescription with those reported. In this case,a combination of chemical composition analysis and network pharmacology is necessary to improve the prediction accuracy of compound activation in TCM Formulae 16-17.Therefore, the chemical analysis by UPLC-QTOF/MS and interactive network pharmacology was taken as efficacious tool for the dissecting and indicting for the pharmacologic mechanisms of XQCF.
In this study, a UPLC-QTOF/MS analytic technique was processed firstly for the identification of the compounds in XQCF. Then the compounds database was established for the perdition of targets. Candidate targets were checked from several database and mapped to predicted therapeutic targets in XQCF. Several networks were constructed to illustrate the complex relationship amongst compounds, targets and proteins. Finally,potential targets, related biological process and pathways were correspondingly explored for further investigations of targets pharmacological mechanisms. Furthermore,the targets contribution and their distribution were evaluated based on efficacy-oriented compatibility conformation of XQCF.
Materials and Methods
Compounds identification by UPLC-QTOF/MS
Chemicals and reagentsUPLC-MS chromatography-grade Acetonitrile and methanol were purchased from Fisher Chemical Company (Geel,Belgium). Formic acid was purchased from Sigma-Aldrich. Ultrapure water was purified by a Millipore water purification system (Millipore, Billerica,MA, USA). Other reagents of analytical grade were purchased from Guangzhou Chemical Reagent Factory(Guangzhou,China).
Yiyiren,Daoya,Shanzha,Danzhuye,Gouteng,Chantui and Gancao were purchased from Guangzhou Materials Medica Co.Ltd., and identified by Professor Hai-bo Huang, Department of Pharmacognosy, Guangzhou University of Chinese Medicine.Voucher specimens were preserved in Institute of Clinical Pharmacology,Guangzhou University of Chinese Medicine, Guangzhou,China.
Preparation of XQCF was according to China Pharmacopoeia (2015 edition) and modified as the follows: (1) 89 g of Yiyiren (YYR) and 89 g of Daoya(DY)were mixed evenly,extracted with water(1:10,w/v)at 100℃twice(2h for each time).The combined filtrated solution was concentrated at 55 ℃ to a condensed solution with relative density of 1.10mg/mL, and then,added ethanol to bring the final ethanol concentration to 45%. After being stood for 24 h, the supernatant was recycled ethanol under a vacuum to obtain extraction of Yiyiren and Daoya germinates. (2) The homogeneous mixture of the rest five herbal materials as 45 g of Shanzha, 67 g of Danzhuye 34 g of Gouteng, 11g of Chantui and 11 g of Gancao was extracted with water(1:10, w/v) at 100 ℃ twice (2 h for each time), and the combined filtration was concentrated. (3) The combined concentrated solution of extraction of Yiyiren and Daoya germinates and concentrated solution of the rest five herbal materials were mixed evenly,and freezing dried to obtain XQCF sample.
7.4 mg of the XQCF sample was added in 1 mL 15%acetonitrile,vortexed for 3 min and centrifuged at 14,000 rpm for 15 min. The super natant was filtered through0.22μm microfiltration membranes before analysis. 5 μL of the sample were injected into the UHPLC–MS/MS system.
UPLC-QTOF/MS analysis of XQCFThe UPLC-QTOF/MS was performed on a Shimadzu UHPLC system (Shimadzu, Kyoto, Japan) coupled with AB SCIEX Triple TOF 5600+mass spectrometer(AB SCIEX,Foster City, CA, USA) through an electro-spray ionization(ESI)interface.The samples were separated on an Agilent C18 column (3.0 × 50 mm, 2.7 μm, Agilent Technologies Inc., USA). The column temperature was 25°C,and the auto sampler was set at 4 °C.The injection volume was 2 μL. The mobile phase consisting of (A)acetonitrile and (B) water containing 0.1% formic acid was applied in a gradient elution of 20%–70%A at 0–12 min.
The analysis was carried out by ESI source in positive and negative mode with the ion spray voltage in±4,500 V.The MS spectra mass range was m/z 100~1000 Da, and the MS/MS spectra was from m/z 50~1000Da. The collision energy was ±45V and the declustering potential was ±100V. The nebulizer gas and the heater gas were both in 55 psi. The turbo spray temperature was 550 °C.All the collection and analysis of data were processed with the Analyst SoftwareTM2.2(AB SCIEX,Foster City,CA,USA).
Compounds and disease targets database construction
Targets of compoundsOn the basis of the built compounds database, the SMILES strings and 3D molecular structures of the compounds were acquired from PubChem (https://pubchem.ncbi.nlm.nih.gov/), an official chemical database which contains comprehensive chemical information. Then we imported the SMILES strings into TCMSP (http://lsp.nwu.edu.cn/tcmsp.php)18-19, Swiss Target Prediction(http://www.swisstargetprediction.ch/) 20-21 and STITCH database (http://stitch.embl.de/) 22-23 and searched potential targets of the compounds. SWISS and STITCH have the function of predicting targets of small molecules,with the combination of 2D and 3D similarity measures, the inquiry molecules were compared to the library of compounds active on targets. The probability score in SWISS and confidence score in STITCH were set over 0.7 and 0.4 respectively for the targets predicted.All the targets were predicted based on the organism of“homo sapiens”.
Targets of IFDXQCF has the efficacy of treating indigestion. The targets were therefore searched with the keyword including “functional dyspepsia”, “indigestion”and “gastrointestinal disorder”. The human genes related to indigestion were obtained from several online databases. Online Mendelian Inheritance in Man 24(OMIM, https://omim.org/) is an authoritative online compendium with continuously updated catalog of human genes and genetic disorders.Therapeutic Target Database 25 (TTD, https://db.idrblab.org/ttd/) provide comprehensive information with fully referenced including targeted disease,therapeutic protein and nucleic acid targets, pathway information and the corresponding drugs directed at each of these targets. GeneCards(http://www.genecards.org/) 26 is a database aims at quick searching of biomedical information about genes.The encoded proteins, relevant diseases and functional information are provided. Likewise, ‘Homo sapiens’proteins related to the disease were selected.
Finally, all the targets in the database were queried in UniProt KB to obtain standard targets’ names. Then the duplicated genes were removed from those databases and the corresponding targets were assembled for the IFD targets database.
Interactive network construction and data mining
Network ConstructionOn the basis of target database,gene symbols of the IFD targets in XQCF were uploaded onto the String Database(https://string-db.org)to acquire protein-protein interaction (PPI) data. String Database collects and presents known and predicted PPI with a confidence score and accessory information. The confidence score represents the interaction positive relationship of the protein. Only protein interactions with the confidence score 0.400 were kept for further study.The interaction between portions was evaluated using the online tool named Multiple Proteins of String, and the results was acquired and saved as TSV document format as prophase data.
To achieve a systematic understanding of the complex relationships among compounds, targets, and diseases,network visualization software Cytoscape(http://cytoscape.org/, ver. 3.2.1) was used to build all networks.The selected targets and data were inputted into Cytoscape to construct the following 3 networks: (1)compound-compound target network; (2)compound-therapeutic target network;(3)PPI network of IFD targets in XQCF.
Gene Ontology Enrichment and Pathway Analysis.
The Gene Ontology(GO)enrichment analysis and KEGG Pathway Analysis was dissected via ClueGo, a comprehensive set of functional annotation tools for the investigators of biological meaning and mechanisms behind large lists of genes. The cluster of GO biological process enrichment analysis was set to 3, the Kappascore was 0.25, the percent of Genes was 10% and the level was 0~3.The cluster of KEGG Pathway Analysis was set to 2, the Kappascore was 0.25 and the percent of Genes was 5%.
Results
Q-TOF-MS analysis of compounds in XQCF
The XQCF concentrate sample was analyzed by UPLC-Q-TOF-MS. A total of 53 compounds were identified or tentatively characterized, including 23 favonoids, 11 alkaloids, 8 phenolic acids, and 11 miscellaneous compounds including 3 organic acids, 3 amino acids, 2 triterpenoid saponins, 2 saccharides and 1 coumarin(Figure 1,Table 1).
Identification of Flavones their glycosides in XQCF Flavones and isoflavone.A total of 10 flavones and 13 of flavonoid glycosides were identified from the samples and most of the compounds were devoted from Shanzha,Yiyiren, Danzhuye and Gancao. Characteristic fragment ions origin from a Retro-Diels-Alder (RDA) cleavage 27 together with continuous or coinstantaneous losses of CO,CH3, H2O, OH and CO2 are the identification evidence of this type of compounds 28. Compounds 9, 11, 24, 49,50, 51 and 52 were flavones and elucidated as quercetin,liquiritigenin, luteolin, kaempferol, diosmetin, naringenin and tricin,respectively.Compound 24 was a flavone with basic skeleton of flavones and was identified as luteolin.It exhibited the ions at m/z 287.0539 in the ESI positive ion mode and generated product ions at m/z 153.0170(1,3A-) and 135.0435(1,3B-) science C-ring underwent a RDA fragmentation.
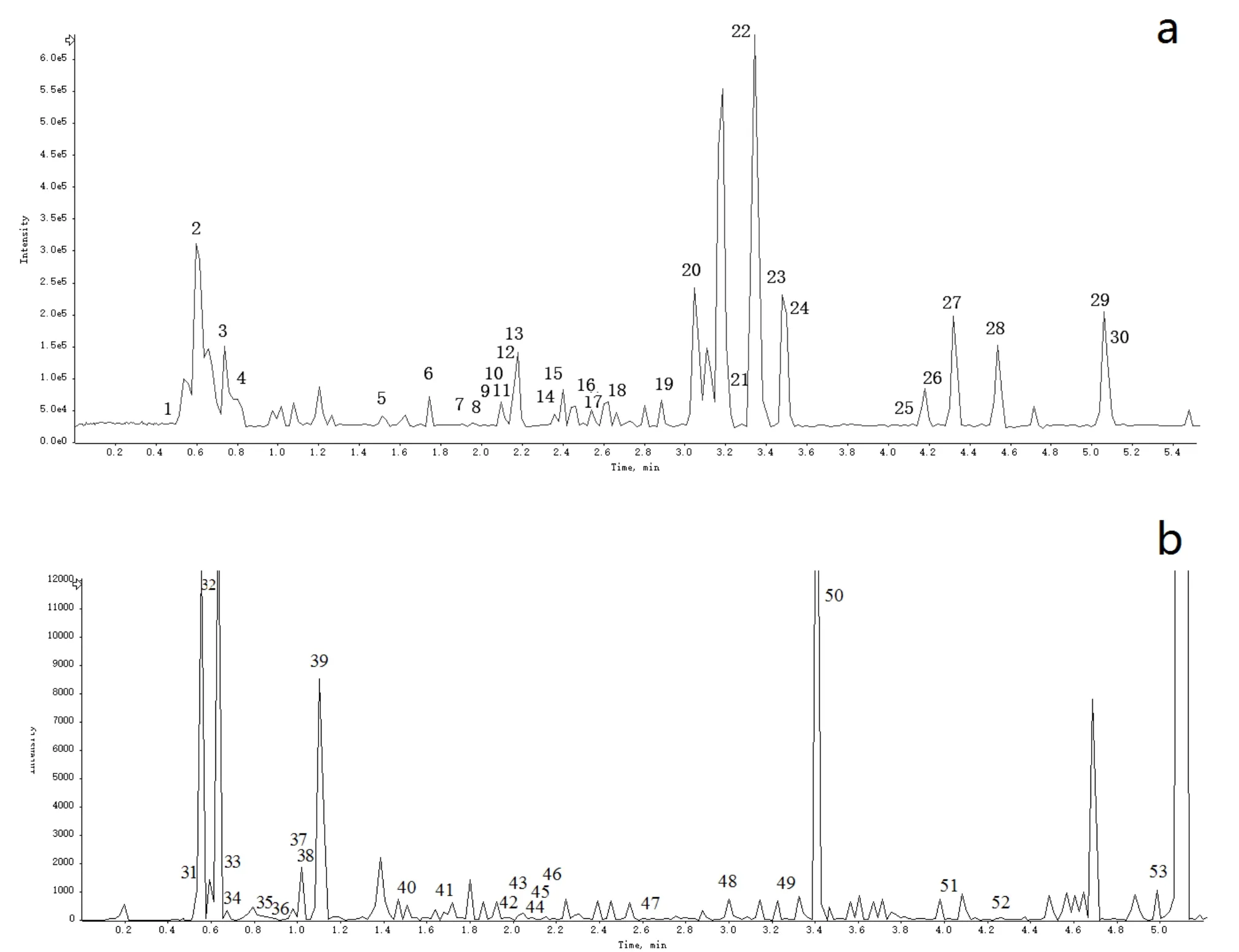
Figure 1 Representative base peak chromatogram in UPLCQ-TOF-MS analysis of XQTF(a)Chromatograms in positive ion mode.(b)Chromatograms in negative ion mode.

*SZ:Shanzha;DY:Daoya;YYR:Yiyiren,DZY:Danzhuye;GT:Gouteng;CT:Chantui;GC:Gancao.
Moreover, Compound 53 was characterized as formononetin, an isoflavone in XQCF. It generated the precursor [M+H] - ions at m/z 267.0666. With the losses of CH3, CO and CO2, product fragment ions at m/z 252.0425 [M-H-CH3] –, 223.0402 [M-H-CO2] – and 195.0452 [MH-CO2-CO] were observed in MS/MS spectra. And then cleavage of the ion at m/z 252.0425 gave rise to fragments at 135.0087(1,3A-) and 132.0271(1,3B-).
Chalcone.In light of the structural characteristics and typical fragment ions of flavanone and chalcone, they showed similar fragmentation pathways science R10=OH could easily transform to the corresponding flavanone isomers 29. Compound 21 and 23 were assigned as licochalcone B and isoliquiritigenin for the type of chalcone.Compound 21 showed [M+H]+ions at m/z 287.0906 and produced predominant fragment ions at m/z 151.0386[M+H-C7H5O2-CH3] + and 193.0502[M+H-C6H5-OH] +, which was the fragmentation behaviors were similar to those of licochalcone B. Meanwhile, compound 23 was also identified as isoliquiritigenin based on those cleavage patterns
Flavonoid glycoside.In addition to the common cleavage pathway of flavone, flavonoid glycoside undergoes typical successive or simultaneous losses of glycosyl segment as well. For instance, compounds 46 and 48 both showed [M-H] – ion at m/z 417. Besides,corresponding fragment [M-H-Glu] –,[M-H-Glu-OH-C8H7] – and [M-H-Glu-C7H3O2] – were observed, which showed ion at m/z 255, 135 and 119 respectively. Accordingly, compounds 46 and 48 were identified as isoliquiritin and liquiritin by comparing with the reference compound and their retention time30Compound 45 showed [M-H] + ion at m/z 549.1622, and fragment ions at m/z 255.0673 corresponding to [M+H-Glu-Api] +. It could be liquiritinapioside which derived from liquiritin with an additional apiosyl substitution. Analogically, other 10 flavonoid glycosides were tentatively defined this on the bases of these cleavage patterns and related literatures,including isoorientin (6), vitexin 2''-O-rhamnoside (7),hyperoside (10), swertisin (12), diosmin (17), orientin(41), rutin (42), vitexin (43), isoquercitrin (44), and isovitexin(47).
Identification of Alkaloids in XQCFAlkaloids in
XRQXC came from GT and only showed in positive ion mode.It’s obvious that compounds 15,18,19,20,22 and 26 had similar fragmentation patterns which indicated that they share common skeleton. Compounds 15, 20, 22 and 26 presented the same [M+H] + ion at m/z 385. In addition, compounds 20 and 22 shared fragment ions at m/z 269.1639 [M+H-C5H8O3] + and 160.0748[M+H-C5H8O3-C5H6N- C2H5] +, the fragmentation behaviors were similar to those of rhynchophylline and isorhynchophylline31. With regard to compounds 15 and 26,which had sharedquasi-molecular[M+H]+ion at m/z 353.1853 [M+H-C5H3O-H] + and 241.1329[M+H-C5H7O3-C2H5] +, were identified as corynoxine B and corynoxine. Therefore, taking the order of the retention times of UPLC chromatograms as reference,compounds 15, 20, 22 and 26 were tentatively identified as corynoxine B, rhynchophylline, isorhynchophylline and corynoxine. Additionally, Compound 18 and 19 showed the same[M+H]+ion at m/z 283,suggesting the incorporation of an additional hydrogen atom into the structure of rhynchophylline or isorhynchophylline.Hence they were identified as isocorynoxeine and corynoxeine according to the retention times as well.Similar situation were applied to other two kinds of isomers including cadambine (14) and 3α-dihydrocadambine (16) along with hirsuteine (27),hirsutine(28)and geissoschizinemethylether(25).
Identification of Phenolic Acids in XQCFThe negative ion mode was much more suitable for phenolic acids analysis and eight compounds of this type were identified.Successive or simultaneous neutral losses of COOH, OH and H are the characteristic behavior of this type of compounds. Compounds 5, 8, 35, 36, 37, 38, 39 and 40 were inferentially identified as cis-p-coumaric acid,p-coumaric acid, cryptochlorogenic acid, neochlorogenic acid, protocatechuic acid, vanillic acid, chlorogenic acid and caffeic acid. For example, compounds 35, 36 and 39 exhibited [M?H] – ions at m/z 353.0876, 353.0872 and 353.0881 with molecular formulae speculated as C16H18O9. Meanwhile, the specific fragment ions of chlorogenic acid32 at m/z 191.0562 [M-H-C9H4O-2OH]– , 135.0453 [M-H-2COOH-C6H5-3OH] – and 173.0455 [M-H-C9H5O-3OH] – were displayed in MS/MS fragment ions, which indicated that they should be isomers of chlorogenic acid. By comparing retention time of the compounds in UPLC system, the compounds were identified as cryptochlorogenic acid (35),neochlorogenic acid (36) and chlorogenic acid (39)respectively.
Other Types of Miscellaneous Compounds in XQCF
Two saccharides were tentatively identified by comparison with authentic references. Compound 1 gave an [M-H] – ion at m/z 179.0565 and showed fragment ions at m/z 89.0254 [M-H-C3H3-3OH] – and 59.0167[M-H-C4H5O-3OH] –, putatively identified as maltose. Compound 31 exhibited [M?H] – ions at m/z 342.1376, the diagnostic fragment ions at m/z 145.0495 by lose of glucose and dehydrogenation reaction, and fragment ions at m/z 85.0295 by ring-opening reaction,which proved to be galactose 33.
Three amino acids were analyzed in positive ion mode including valine, isoleucine and phenylalanine. The mass spectra of Compound 2 exhibited a molecular ion [M+H]+ at m/z 118.0859 and fragment ions at m/z 72.0839[M+H-COOH-H] + and 58.0696 [M+H-COOH-CH3] +,which proved to be valine. Similarity, compound 3 showed [M+H] + ions at m/z 132.1016 and MS/MS fragment ions at m/z 86.0983 [M+H-COOH-H] + and 58.0528 [M+H-COOH-C2H5] +, the fragmentation behaviors were similar to those of isoleucine.
Two triterpenoid saponins which were accurately identified in positive ion mode by compared with references as glycyrrhizic acid and glycyrrhetinic acid,are the individual components of Gancao 34. Compound 29 displayed the[M+H] +at m/z 823.4086,and the yield ion at m/z 647.3764 [M+H-C6H8O6] + and 471.3445[M+H-2C6H8O6] +was similar to the losses of glycyrrhizic acid. Compound 30, which was identified as glycyrrhetinic acid, showed [M+H] + at m/z 471.3455,and the fragment ions at m/z 235.1682[M+H-OH-4CH3-CO-C10H11] + and 317.2212[M+H-OH-3CH3-C7H8]+.
Three organic acids were identified by comparison with authentic references in negative ion mode.Compound 32 had a molecular ion [M?H] – at m/z 191.0560,and the MS/MS fragment ions at m/z 127.0404 and 87.0106 corresponded to [M-COOH-OH-2H] – and[M-H-C4H5-3OH] –, putatively identified as quinic acid.The MS chromatograms of compound 33 exhibited a quasi-molecular ion at m/z 133.0145 [M?H] –, and two diagnostic fragment ions at m/z 115.0040[M-H-H2O] –,72.9947 [M-H-COOH-CH3] –and 71.0158[M-H-COOH-OH] –, which proved to be malic acid.Compound 34 exhibited [M?H] – ions at m/z 191.0205 and fragment ions at m/z 111.0097 corresponded to[M-H-CO2-2H2O] –, which was identified as citric acid35.
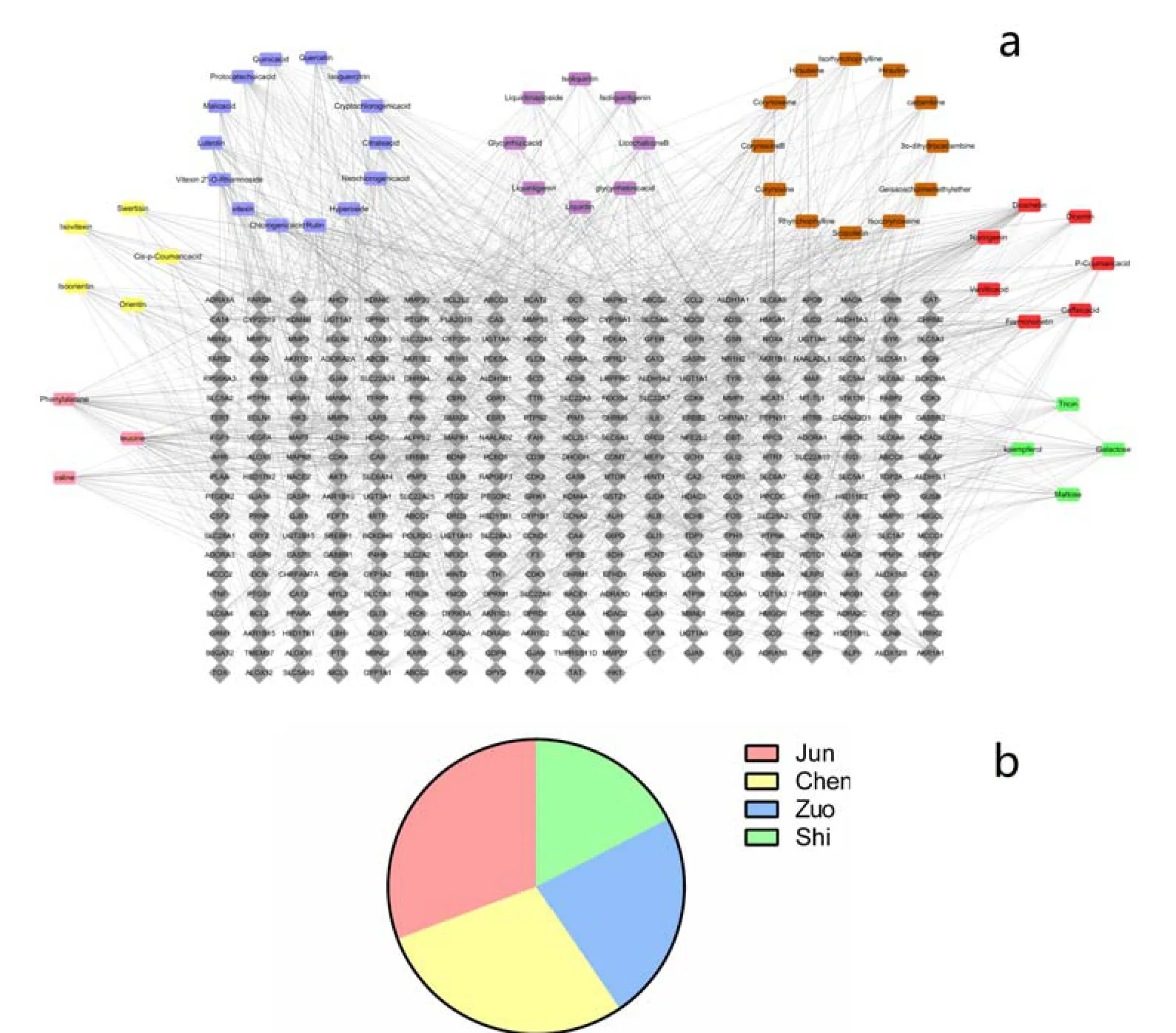
Figure 2 Network and distribution of compound targets in XQTF
Targets prediction and screening
Targets identification of XQCF and IFDChinese Herbal Compounds take curative effect via multiple components acting on multiple targets. In the present work, the compounds were classified into 7 herbs attribution group in the form of compound-herb according to TCMSP database, and several compounds coexisted in groups as shared active ingredients of each herb in XQCF. The shared active ingredients demonstrated the multiple interactions between compounds and herbs. Afterwards, the targets of each compound were screened. Within the limits of prescriptive score, the displayed 15 targets from SWISS and top 20 from STITCH were selected. After deleting duplicate targets of the same compound between the two databases, a total of 373 targets interacted with 53 active ingredients in XQCF were predicted as compound-target dataset. As shown in Figure 2a, 373 targets were identified for 53 compounds of XQCF on the basis of herbs attribution groups. The distribution ratio of the targets from herbs of Jun, Chen, Zuo and Shi were 92:86:69:52, which was expressed as percentages in Figure 2b. Meanwhile, targets of IFD were obtained as disease-target dataset from various databases including OMMI and TTD by the keywords of “functional dyspepsia”,“indigestion”and“gastrointestinal disorder”.
Therapeutic targets in XQCF in the treatment of IFD
The predicted targets of the active ingredients in XQCF were mapped to the targets of IFD to get shared targets,which were regarded as candidate targets of IFD.Among them, 64Therapeutic targets with no repeats were acquired. In this study, 64 Therapeutic targets were identified for 53 compounds of XQCF (Figure 3a). And the proportion of therapeutic targets from in Jun, Chen,Zuo and Shi herb couple were 39:34:24:10, as it shown graphically in Figure 3b.
Network construction and analysis
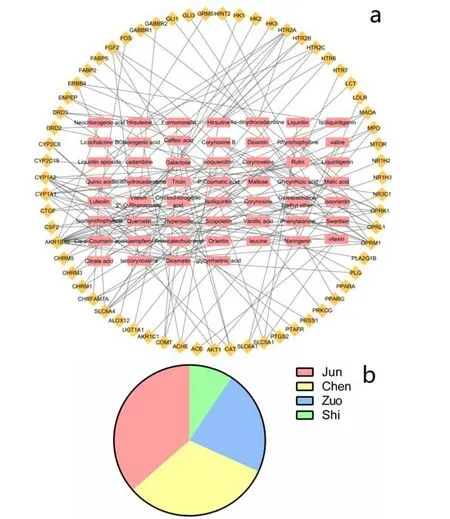
Figure 3.Network and distribution of therapeutic targets in XQTF
Compound- compound target network and compound- disease target network Thecompound-compound target network was composed of 327 edges and 373 nodes, in which 13 compound nodes and 373 compound target nodes were included, as shown in Figure 2. As it shown in the network, a single target can be adjusted by different active components and the same component can correspond to different targets,which suggests the treatment characteristics of multi-component and multi-target of XQCF. From this network, a preliminary observation of the relationship between active compounds and compound targets was exhibited.
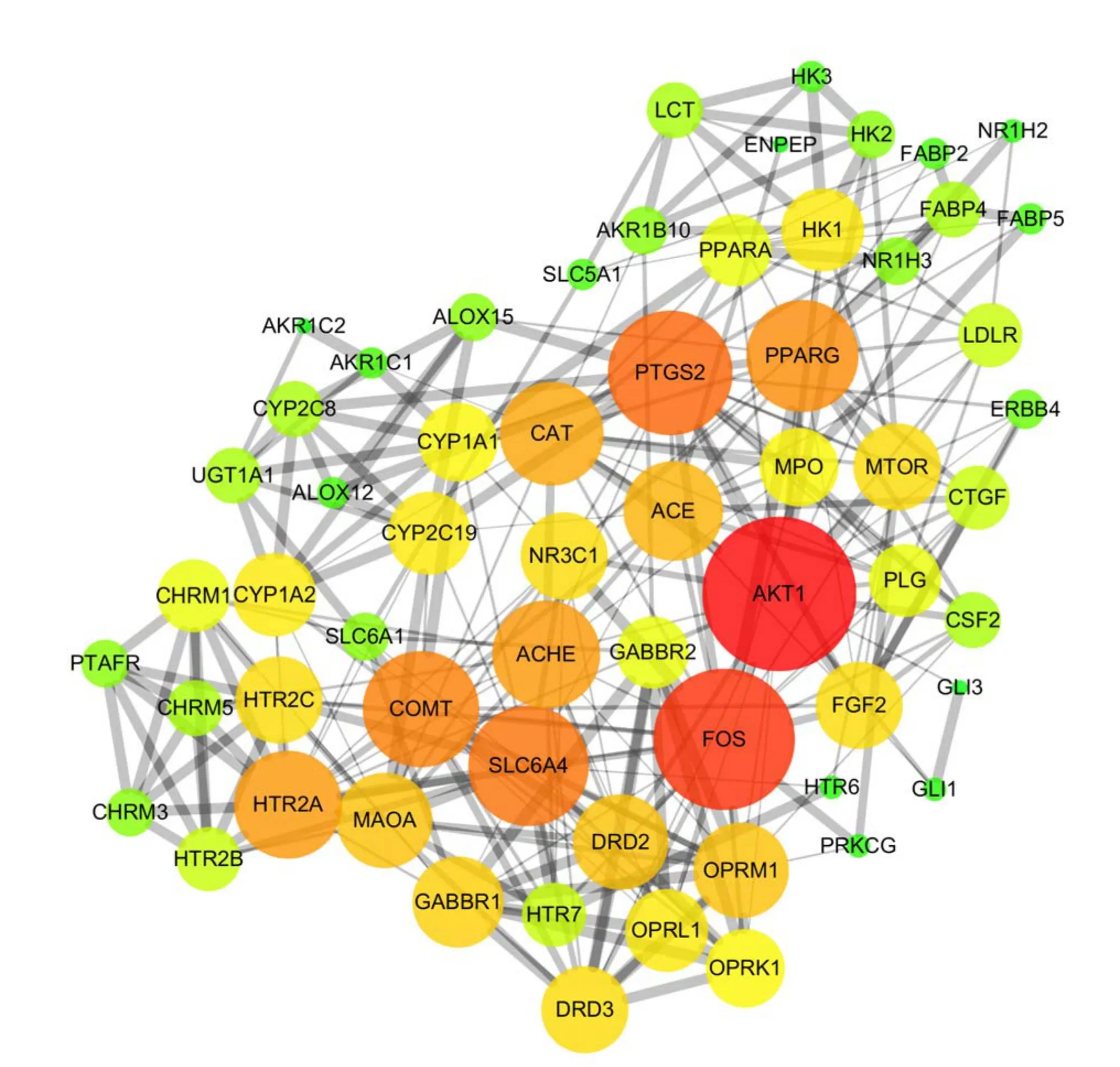
Fig 4 PPI Network
For the prediction of key compounds for the treatment of IFD by XRQXC, a compound-disease target network was performed on the basis of the therapeutic targets dataset.The network consisted of 64 nodes and 80 edges,including 53 compounds and 64 therapeutic targets,and it indicated the compounds and targets which were closely related to the development of the disease, as shown in Figure 3. The analysis result of network showed that the compounds having high degree value over 5 were considered as potential key active compounds, including galactose, isocorynoxeine, corynoxine, hirsutine,diosmetin, quercetin and vitexin 2''-O-rhamnoside.Moreover, the top 5 therapeutic targets exhibiting high degree value were AKR1B10, CYP1A2, CYP1A1,SLC6A4, OPRM1, OPRL1 and OPRK1. However, the deeper interaction between targets during the treatment bioprocess also played a significant role. Therefor a PPI network is necessary for the further exploration.Protein-protein interactions (PPI) network of IFD targets in XQCF PPI network reveals the underlying pharmacological mechanisms of TCM Formulae against disease. In this study, the PPI network was constructed using Cytoscape by importing the data of node1, node2 and combined score in the TSV document from String database. The network was composed by 68 nodes(therapeutic targets)and 474 edges(Figure 4,Table 2).As Figure 4 suggests, the node size and color reflected degree value, a bigger node size meant a higher value.And with the increasing of degree value, the node circle was turned from green to red. Moreover, the thickness of the edge reflected the value of combine score, and a higher score was represented by a thicker line. The PPI network indicated that there were 26 targets with degree over 15,and the top 5 with higher degree including AKT1,FOS, PTGS2, SLC6A4 and COMT could be considered as vital targets in the pharmacological mechanisms of XQCF.GO biological process enrichment and KEGG Pathway enrichment analysis For further investigations of targets pharmacological mechanisms, the GO biological process and KEGG Pathway enrichment were carried out by ClueGO function module of Cytoscape.The parameter “P-Value” of enrichment result means statistical significance of the GO term and lower P-Value implies higher significance, which means a more reliable prediction. Figure 5a revealed 10 main GO biological processes referred in the treatment of IFD. Table 3 indicated possible major biological processes with low P-Value including digestion, digestive system process,positive regulation of lipid biosynthetic process and serotonin receptor signaling pathway. Meanwhile, KEGG pathway result suggested that there were 6 KEGG pathways observed highly correlative with pharmacological biological behaviors,as shown in Figure 5b. And pathways in Table 4 such as Neuroactive ligand-receptor interaction, Serotonergic synapse,Calcium signaling pathway, Carbohydrate digestion and absorption with low P-Value also revealed possible mechanisms involved from a different perspective. The therapeutic targets distribution of Jun,Chen, Zuo and Shi herb couple were also calculated based on those momentous biological processes and pathways.The result showed that the specific value was 68:56:36:11,as shown in Figure 5c.
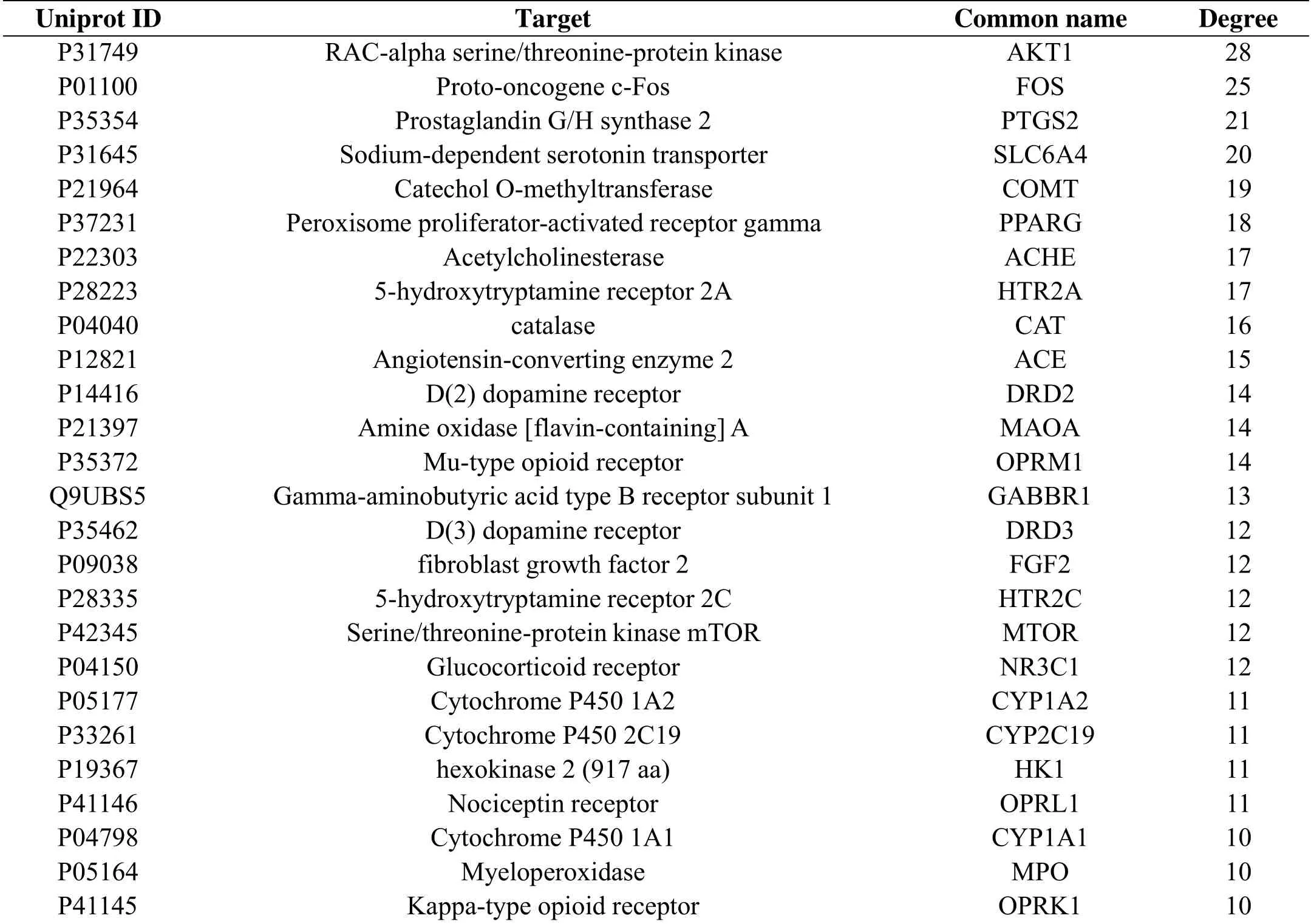
Table 2 Significant therapeutic targets with degree over 10 in PPI network
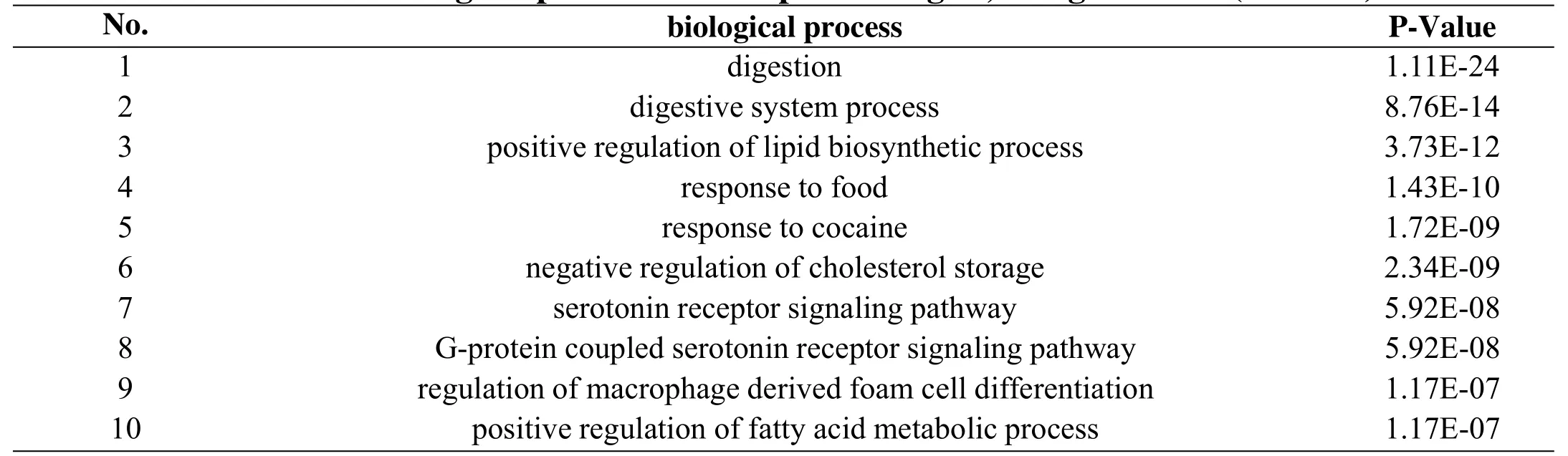
Table 3 GO biological process of therapeutic targets,using ClueGO(P<0.05)
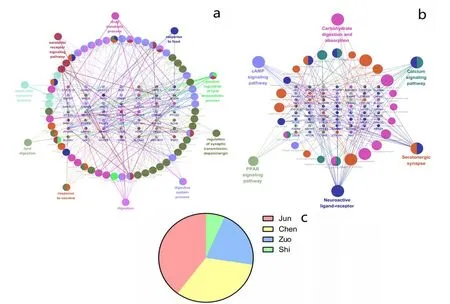
Figure 5 GO biological process and KEGG pathway analysis

Table 4 KEGG pathway analysis of therapeutic targets,using ClueGO(P<0.05)
GO biological process analysis showed that XQCF had regulating functions on digestive process, digestive system function and lipid metabolism.And the regulation of metabolism and response of neuroreceptor was also included,which illustrated that the treatment process may have important links with the brain-gut axis. The GO biological process network also proved the mediation on the processes in cells, tissues, organs, metabolism, and immunity. All the above indicated that the regulation of multiple processes by XQCF was significant in the treatment of IFD for its complex nosogenesis.Meanwhile,KEGG pathway network showed that the interaction of targets connected several important pathways closely,including neuroactive ligand-receptor interaction,serotonergic synapse, calcium signaling pathway,Carbohydrate digestion and absorption and cAMP signaling pathway. Signaling molecules such as 5-HT,cAmp, Ca+ were shared and activated inhibited the pathways simultaneously. The pathways had obviously relativity with absorption and digestion of foods, which was the primary process to regulate in the treatment of IFD.
Discussion
Prescription compatibility is the core of TCM Formulae for the sake of better curative efficacies and fewer side effects 36.In TCM,it follows the rule of“Jun,Chen,Zuo and Shi” which corresponding to the role of “monarch,minister, assistant, and messenger”. Jun herbs are the main efficacy-contributing herbs in a TCM Formulae and play the role of monarch.Chen herbs mean adjuvant ones to enhance effects of Jun herbs and aim at accompanying symptoms simultaneously. Zuo herbs usually served to enhance the effect of Jun and Chen herbs and alleviate adverse effects of the Formulae as assistant. Shi herbs refer to coordinating the drug actions of the prescription as a messenger 37. In XQCF, SZ and DY are defined as Jun herbs in this Formulae with the effects of stimulating appetite and digestion, clearing heat and relieving restlessness. Chen herbs are YYR and DZY, which existing the effects of invigorating spleen and stomach,dehumidification and diuresis.As Zuo herbs, GT and CT serve as assistant to calm endogenous wind and help sedation. GC is the Shi herb with function of nourishing and harmonizes the effects of the whole Formulae.
In this study, the position of herbs and curative effect contribution were analyzed based on distribution of the number of targets. In the compound-target dataset,therapeutic targets dataset and biological process and pathway involved targets dataset, the targets in each research module were classified into 4 groups according to their compounds and herbs source, namely Jun, Chen,Zuo and Shi. As compound-target dataset statistics showed, the number of targets of Jun, Chen and Zuo group tended to be approximately equal and the number from Shi group was slightly smaller. However, when the predicted targets of the active compounds in XQCF were mapped to the targets of IFD and shared targets were selected as therapeutic targets, the difference between each group was obviously changed as the ratio of Jun and Chen group were raised while Zuo and Shi were reduced.Moreover, the sequence of the ratio was: Jun, Chen, Zuo and Shi, which was conformed to the rule of prescription compatibility in TCM. With regard to the therapeutic targets in the significant biological processes and pathways relating to the treatment of IFD, the regularities of distribution was more reasonably agreed with the contribution of Jun, Chen, Zuo and Shi. The ratio of Jun and Chen were further increased and ratio of Shi was decreased again. Our study proved that compatibility in TCM was significant to the effectiveness and uniqueness of the Formulae. The efficacy-oriented compatibility was confirmed based on the therapeutic targets mapping changing the ratio of Jun,Chen,Zuo and Shi group.
In the PPI network, the importance of each node is evaluated by the betweenness centrality(BC) and Degree Centrality(DC)38-39.In this study,DC was chosen as an important parameter for the identification and evaluation of essential proteins.For achieving the reliable prediction,targets with high degree were analyzed for their possibly leading role in the process of pharmacological effects for IFD. AKT1, namely RAC-alpha serine/threonine-protein kinase, is one of the kinase of AKT family members and expressed ubiquitously to mediate multiple processes through a series of downstream substrates.AKT is known to be a downstream effector of PI3K, and the phosphorylated AKT could activate PI3K/AKT signal pathway which counts for the protective effect on small intestine 40. Moreover, the other study disclosed that AKT exert substantial physiological functions and to regulate several intracellular effects of epidermal growth factor (EGF)41, which is a vital peptide growth factor exerting biological action in gut. Studies performed AKT could play a functional role in mediation of gastric acid section42 through regulation from EGF on H+/K+-ATPase gene expression43. Meanwhile, Akt1 is known to be closely associated with stimulation of gastric production44. With the function on transforming protein into proteoses and peptones, gastric acid potently promotes digestion process45 and regulating gut acid takes a leading role in the treatment of dyspepsia.Fos(c-Fos) is a marker of stimulus-induced neural activation.As the neuronal and neuronal from alimentary system were integrated in the process of ingestion46, the ingestion of a large satiating meal contributed to c-Fos expression of hindbrain, while the expression was low after ingestion of a rationed meal or no meal47.Furthermore, a liquid nutrient meal test indicated that the Fos intimately relevant to IFD reduced by crapulence48.SLC6A4, a sodium-dependent serotonin transporter with the alternate name of serotonin transporter protein(SERT),is an important signaling molecule affecting digestion system motor and sensory functions.Ninety-five percent of the body’s serotonin is synthesized in enterochromaffin cells in the gut49.In the pathological state of IFD,SLC6A4 had a higher expression and HTR7 existed lower tendency. Meanwhile, exogenous 5-HT induced duodenal ion transport was abnormal in IFD,which indicated the involvement of 5-HT in IFD pathogenesis50. It has been approved that SLC6A4 had high affinity with 5-HT and can intake 5-HT in the intercellular space then regulate signal transduction.Therefor the expression of SLC6A4 has deep interaction with the secretion of 5-HT51and its variform receptor,including the predicted targets of HTR2, HTR5, HTR6 and HTR7 in this study.Impaired 5-HT receptors such as HTR7 mediated duodenal epithelial transport or signaling and impacted on pathology in IFD. Furthermore, IFD patients had lower expression of HTR752.With regard to COMT, an important enzyme in brain gut axis and pain sensitivity53,influenced the susceptibility of dyspepsia to an extent54.Accordingly, a series of targets in this study with function of simultaneously manipulation associated with physiological process and mechanisms of IFD closely. Consequently, targets above could be considered as therapeutic targets relate to pharmacological mechanisms of XQCF.
In the analysis of GO biological process and KEGG pathway,pathways and process which involved in central nervous system(CNS),enteric nervous system(ENS)and autonomic nervous system (ANS) displayed high relevance with remedial mechanisms. Those highly correspond to the regulatory structure named brain-gut axis, the signal system which regulates digestion process55-56. Furthermore, brain-gut peptides (BGP)implicates in the bidirectional gut–brain communication57, and its alteration affects the bidirectional interaction between nervous system and gut in IIFD58 Accordingly,meditating the mRNA expression of neuropeptides via pathways including neuroactive ligand-receptor interaction and serotonergic synapse,with of regulation of pathways with relate to digestion system simultaneously such as Carbohydrate digestion and absorption, Calcium signaling pathway, cAMP signaling pathway. All this leads up to the inference that pharmacologic mechanisms of XQCF on IIFD correlated with the predicted targets and pathways related in brain–gut axis to a great extent.
Conclusions
TCM prescription has been extensively applied clinical and plays an indispensable role for the treatment of IFD.This study identified the compounds from XQCF by UPLC-QTOF/MS for the first time and 53 active ingredients were attributed. The many-many relationship between multiple ingredients and multiple targets were predicted and the mechanisms of XQCF against IFD were explored network pharmacology. Our study found that brain–gut axis could correlate with IFD closely through several important targets and pathways. The study not only provided the experimental and theoretical basis for further research of XQCF, but also illustrated a reasonable method worth intensive study on pharmacodynamic mechanisms of TCM Formulae.
1. Ganesh M. Functional dyspepsia in children.Pediatric annals 2014,43:101-105.
2. De Giacomo C, Valdambrini V, Lizzoli F, et al. A population‐based survey on gastrointestinal tract symptoms and Helicobacter pylori infection in children and adolescents. Helicobacter 2002, 7:356-363.
3. Chen J D Z, Lin X, Zhang M, et al. Gastric myoelectrical activity in healthy children and children with functional dyspepsia.Dig Dis Sci 1998,43:2384-2391.
4. Polivanova T V, Smirnova O V, Gorbacheva N N, et al. Efficiency of Very Low Doses of Antibodies to S100 Protein in the Complex Therapy of Functional Dyspepsia in Children. Bull Exp Biol Med 2015,159(3):341-343.
5. Bi X,Gong M.Review on prescription compatibility of shaoyaogancao decoction and reflection on pharmacokinetic compatibility mechanism of traditional chinesemedicine prescription based on in vivo drug interaction of main efficacious components.Evid Based Complement Alternat Med:eCAM 2014,2014:208129.
6. Cui DN, WangX, ChenJQ, et al. Quantitative Evaluation of the Compatibility Effects of Huangqin Decoction on the Treatment of Irinotecan-Induced Gastrointestinal Toxicity Using Untargeted Metabolomics.Front Pharmacol 2017,8:211.
7. Teschke R, Wolff A, Frenzel C, et al. Herbal traditional Chinese medicine and its evidence base in gastrointestinal disorders. World J Gastroenterol 2015,21(15):4466-4490.
8. Wang X, Huang X, Zhao X, et al. Prokinetic and laxative effects of Xiao'erQixingcha, a household pediatric herbal Formula. Afr J Tradit Complement Altern Med 2015,12(5):89-98.
9. Committee C P. Pharmacopoeia of the People's Republic of China.Part 2010,1:392-3.
10. Qu C, Yang G H, Zheng R B, et al. The immune-regulating effect of Xiao’er Qixingcha in constipated mice induced by high-heat and high-protein diet. BMC Complement Alternative Med 2017,17:185.
11. Xiao S, Luo K, Wen X, et al. A pre-classification strategy for identification of compounds in traditional Chinese medicine analogous Formulas by high-performance liquid chromatography–Mass spectrometry. J Pharmd Biomed Anal 2014, 92:82-89.
12. Qi Y,Li S,Pi Z,et al.Chemical profiling of Wu-tou decoction by UPLC–Q-TOF-MS.Talanta 2014, 118:21-29.
13. Hopkins A L. Network pharmacology. Nature Biotechnol 2007,25:1110.
14. SJ Song, RJ Xu, LJ Xiu, X Liu, et al.Network pharmacology-based approach to investigate the mechanisms of YiyiFuziBaijiang Powder in the treatment of malignant tumors.Tradi Med Res 2018,3:295-306.
15. J Hao, SJ Li. Recent advances in network pharmacology applications in Chinese herbal medicine.Tradi Med Res 2018,3:260-272.
16. Ma X,Lv B,Li P,et al.Identification of "Multiple Components-Multiple Targets-Multiple Pathways"Associated with Naoxintong Capsule in the Treatment of Heart Diseases Using UPLC/Q-TOF-MS and Network Pharmacology.Evid Based Complement Alternat Med 2016, 2016:9468087.
17. Liu L, Du B, Zhang H, et al. A network pharmacology approach to explore the mechanisms of Erxian decoction in polycystic ovary syndrome.Chin Med 2018,13:46
18. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines.J Cheminform 2014,6:13.
19. Li P, Chen J, Wang J, et al. Systems pharmacology strategies for drug discovery and combination with applications to cardiovascular diseases. J Ethnopharmacol 2014,151:93-107.
20. Gfeller D,Grosdidier A,Wirth M,et al.Swiss Target Prediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res 2014,42:32-38.
21. Kiefer F, Arnold K, Künzli M, et al. The SWISS-MODEL Repository and associated resources.Nucleic Acids Res 2008,37:387-392.
22. Kuhn M, von Mering C, Campillos M, et al.STITCH: interaction networks of chemicals and proteins.Nucleic Acids Res 2007,36:684-688.
23. Kuhn M, Szklarczyk D, Franceschini A, et al.STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res 2009,38:552-556.
24. Amberger J S, Bocchini C A, Schiettecatte F, et al.OMIM. org: Online Mendelian Inheritance in Man(OMIM?), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2014, 43:789-798.
25. Li Y H, Yu C Y, Li X X, et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res 2017, 46:1121-1127.
26. Rebhan M, Chalifa-Caspi V, Prilusky J, et al.GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 1998, 14:656-664.
27. Wolfender J L, Waridel P, Ndjoko K, et al.Evaluation of Q-TOF-MS/MS and multiple stage IT-MSn for the dereplication of flavonoids and related compounds in crude plant extracts. Anal 2000,28:895-906.
28. Huang P, Ke H, Qiu Y, et al. Systematically Characterizing Chemical Profile and Potential Mechanisms of QingreLidan Decoction Acting on Cholelithiasis by Integrating UHPLC-QTOF-MS and Network Target Analysis. Evid Based Complement Alternat Med 2019,2019.
29. Zhang Y, Wang C, Yang F, et al.UHPLC-ESI-Q-TOF-MS/MS analysis, antioxidant activity combined fingerprints for quality consistency evaluation of compound liquoricetablets.RSC Advances 2018,8:27661-27673.
30. Qi Y,Li S,Pi Z,et al.Chemical profiling of Wu-tou decoction by UPLC–Q-TOF-MS.Talanta 2014, 118:21-29.
31. Xie S,Shi Y,Wang Y,et al.Systematic identification and quantification of tetracyclic monoterpenoidoxindole alkaloids in Uncariarhynchophylla and their fragmentations in Q-TOF-MS spectra.J Pharm Biomed Anal 2013,81:56-64.
32. Wang Y, Wen J, Zheng W, et al. Simultaneous determination of neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid and geniposide in rat plasma by UPLC‐MS/MS and its application to a pharmacokinetic study after administration of Reduning injection. Biomed Chromatogr 2015, 29:68-74.
33. Tüting W, Adden R, Mischnick P. Fragmentation pattern of regioselectively O-methylated maltooligosaccharides in electrospray ionisation-mass spectrometry/collision induced dissociation. International J Mass Spectrom 2004,232:107-115.
34. Wang Y, He S, Cheng X, et al.UPLC–Q-TOF-MS/MS fingerprinting of traditional chineseFormulaSiJunZiTang. J Pharm Biomed Anal 2013,80:24-33.
35. Silva MFGD, Dionísio AP, Abreu FAP, et al.Evaluation of nutritional and chemical composition of yacon syrup using H NMR and UPLC-ESI-Q-TOF-MS. Food Chem 2018, 245:1239-1247.
36. Zhou M,Hong Y,Lin X, et al.Recent pharmaceutical evidence on the compatibility rationality of traditional Chinese medicine. J Ethnopharmacol 2017,206:363-375.
37. Wu L,Wang Y,Li Z,et al. Identifying roles of"Jun-Chen-Zuo-Shi" component herbs of QiShenYiQiFormula in treating acute myocardial ischemia by network pharmacology.Chin Med 2014,9:24.
38. Tang X, Wang J, Zhong J, et al. Predicting essential proteins based on weighted degree centrality.IEEE/ACM Transactions on Computational Biology and Bioinformatics(TCBB)2014,11(2):407-418.
39. Vallabhajosyula R R,Chakravarti D,Lutfeali S,et al.Identifying hubs in protein interaction networks.PloS one 2009,4:5344.
40. CHEN S, CHEN Y, You P, et al. Role of PI3K/Akt signaling in the protective effect of magnesium sulfate against ischemia-perfusion injury of small intestine in rats.Chin Med J 2010,123:1447-1452.
41. Wang X, McCullough KD, Franke TF. Epidermal growth factor receptor-dependent Akt activation by oxidative stress enhances cell survival. J Biochem Physiol 2000,275:14624-14631.
42. Todisco A, Pausawasdi N, Ramamoorthy S, et al.Functional Role of Protein KinaseB/Akt in Gastric Acid Secretion. Mechanisms and Consequences of Proton Transport. Springer, Boston, MA 2002:197-207.
43. Kaise M, Muraoka A, Yamada J, et al. Epidermal growth factor induces H+, K+-ATPase α-subunit gene expression through an element homologous to the 3 ′ half-site of the c-fos serum response element.J Biol Chem 1995,270(31):18637-18642.
44. Ancha H R, Ancha H B, Tedesco D S, et al.Inhibition of epidermal growth factor receptor activation enhances in vivo histamine-stimulated gastric acid secretion in the rat.Dig Dis Sci 2006,51:274-281.
45. Low AG. Nutritional regulation of gastric secretion,digestion and emptying. Nutr Res R 1990, 3:229-252.
46. Olson B R, Freilino M, Hoffman G E, et al. c-Fos expression in rat brain and brainstem nuclei in response to treatments that alter food intake and gastric motility.Mol Cell Neurosci 1993,4:93-106.
47. Rinaman L, Baker E A, Hoffman G E, et al.Medullary c-Fos activation in rats after ingestion of a satiating meal. Am J Physiol Regul Integr Comp Physiol 1998,275:262-268.
48. Winston J H, Aguirre J E, Shi X Z, et al. Impaired interoception in a preclinical model of functional dyspepsia.Dig Dis Sci 2017,62:2327-2337.
49. Kim D Y, Camilleri M. Serotonin: a mediator of the brain–gut connection. Am J Gastroentero 2000, 95:2698.
50. Witte A B, D’Amato M, Poulsen S S, et al.Duodenal epithelial transport in functional dyspepsia:Role of serotonin. World J Gastroenterol Hepatol 2013,4:28.
51. Jess U, El Far O, Kirsch J, et al. Interaction of the C-terminal region of the rat serotonin transporter with MacMARCKS modulates 5-HT uptake regulation by protein kinase C. Biochem Biophys Res Commun 2002,294(2):272-279.
52. Witte A B. The small bowel and functional dyspepsia-peptide hormones and neurotransmitters.Instf?rmedicin, Solna/Dept of Medicine, Solna 2013.
53. Oshima T, Toyoshima F, Nakajima S, et al. Genetic factors for functional dyspepsia. J Gastroenterol Hepatol 2011,26:83-87.
54. Tahara T, Arisawa T, Shibata T, et al. COMT gene val158met polymorphism in patients with dyspeptic symptoms.Hepato-gastroenterol 2008,55:979-982.
55. Mayer E A, Tillisch K. The brain-gut axis in abdominal pain syndromes.Annu Rev Med 2011,62:381-396.
56. Holzer P, Reichmann F, Farzi A. Neuropeptide Y,peptide YY and pancreatic polypeptide in the gut–brain axis.Neuropeptides 2012,46:261-274.
57. Holzer P, Farzi A. Neuropeptides and the microbiota-gut-brain axis. Microbial endocrinology:The microbiota-gut-brain axis in health and disease.Springer,New York,NY 2014:195-219.
58. Huang W, Jiang S M, Jia L, et al. Effect of amitriptyline on gastrointestinal function and brain-gut peptides: a double-blind trial. World J Gastroenterol:WJG 2013,19:4214.
 TMR Modern Herbal Medicine2019年2期
TMR Modern Herbal Medicine2019年2期
- TMR Modern Herbal Medicine的其它文章
- Wenxin granule for cardiac arrhythmia:an overview of systematic reviews
- Terahertz spectral analysis of Yunpian Lurong
- The study on neuroprotective mechanism of water extract of Fomitopsis Pinicola on mesencephala dopaminergic neurons induced by MPP+
- Mechanism analysis of Traditional Chinese Medicine in treatment of Diabetic Nephropathy based on network pharmacology and Traditional Chinese Medicine inheritance support system
- Clinical research progress of ischemic heart failure and prospect of traditional Chinese medicine
