TlMP2 promotes intramuscular fat deposition by regulating the extracellular matrix in chicken
CUl Huan-xian ,LUO Na ,GUO Li-ping,LlU Lu,XlNG Si-yuan,ZHAO Gui-ping ,WEN Jie
State Key Laboratory of Animal Nutrition/Institute of Animal Sciences,Chinese Academy of Agricultural Sciences,Beijing 100193,P.R.China
Abstract The interaction between myocytes and intramuscular adipocytes is a hot scientific topic. Using a co-culture system,this study aims to investigate the regulation of intramuscular fat deposition in chicken muscle tissue through the interaction between myocyte and adipocyte and identify important intermediary regulatory factors. Our proteomics data showed that the protein expression of tissue inhibitor of metalloproteinases 2 (TIMP2) increased significantly in the culture medium of the co-culture system,and the content of lipid droplets was more in the co-culture intramuscular adipocytes. In addition,TIMP2 was significantly upregulated (P<0.01) in muscle tissue of individuals with high intramuscular fat content.Weighted gene co-expression network analysis revealed that TIMP2 was mainly involved in the extracellular matrix receptor interaction signaling pathway and its expression was significantly correlated with triglyceride,intramuscular fat,C14:0,C14:1,C16:0,C16:1,and C18:1n9C levels. Additionally,TIMP2 was co-expressed with various representative genes related to lipid metabolism (such as ADIPOQ,SCD,ELOVL5,ELOVL7,and LPL),as well as certain genes involved in extracellular matrix receptor interaction (such as COL1A2,COL4A2,COL5A1,COL6A1,and COL6A3),which are also significantly upregulated (P<0.05 or P<0.01) in muscle tissue of individuals with high intramuscular fat content.Our findings reveal that TIMP2 promotes intramuscular fat deposition in muscle tissue through the extracellular matrix receptor interaction signaling pathway.
Keywords: TIMP2,ECM,intramuscular adipocytes,muscle satellite cells,chicken
1.lntroduction
Intramuscular fat (IMF) content is the most important evaluation index of chicken meat quality,as it is positively correlated with tenderness and flavor (Gerbenset al.2001;Okeudo and Moss 2005;Fuenteset al.2013;Nogalskiet al.2017). IMF deposition in chicken is regulated by genetic,nutritional,and environmental factors. Studies have shown that there is a direct relationship between skeletal muscle activity and adipose tissue metabolism (Krzysikwalkeret al.2008;Bostr?met al.2012;Liet al.2014,2017;Ojimaet al.2014).Skeletal muscle is an important part of the animal body.Skeletal muscle satellite cells (MSC) are myoblast stem cells located under the basal lamina around skeletal muscle and outside the myofiber plasma membrane.They play an important role in the subsequent growth and regeneration of skeletal muscle (Asakuraet al.2001;Dhawan and Rando 2005;De Coppiet al.2006;Yinet al.2013;Sunet al.2017). Vertebrate muscle cells and adipocytes have been identified to be derived from common mesoderm cells,and are closely related to the regulation of animal growth and development through autocrine and paracrine (Koktaet al.2004;Quinnet al.2005). Moreover,it has also been reported that muscle cells are closely associated with preadipocytes during embryonic development (Ailhaudet al.1992).
The extracellular matrix (ECM) is an important structural component of tissue architecture as an important compound in communicating cells in muscle tissue and plays an important role in fat formation. ECM is mainly composed of two major kinds of macromolecules,namely fibrous proteins and proteoglycans (PGs) secreted by cells into the extracellular mesenchyme (Jones and Jones 2000).The main fibrous protein macromolecules include various glycoproteins (different collagens,fibronectin,and laminin) and elastins (Leeet al.2013). The PGs include those containing one type or a combination of two glycosaminoglycans,such as heparan-chondroitin sulfate PGs (agrin,perlecan,syndecans,glypicans,aggrecan,and some types of collagens) and keratan sulfate (lumican and fibromodulin). The ECM is the substrate for tissue morphogenesis and provides support and flexibility for mature tissues (Jones and Jones 2000). Intracellular signals are transmitted and integrated through different cell surface receptors.ECM-receptor interaction pathways are involved in multiple synthesis pathways of 3 adipose tissues (greater omentum,subcutaneous fat,and organic internal fat),and interact with adipocyte transmembrane receptors,which affect specific adipogenesis (Rajeshet al.2010).It has also been found that ECM interacts with the genes induced by the PPAR signaling pathway. These results indicate that although ECM cannot directly regulate lipid metabolism,it plays an important role in obesity metabolism,thereby affecting fat deposition.
Matrix metalloproteinases (MMPs) are an important enzyme that can degrade ECM,which is composed of a variety of zinc-dependent enzymes. MMPs are increased in adipose tissue of obese individuals to increase matrix degradation and adapt to ECM remodeling caused by fat cell hypertrophy. Adipose tissue can secreteMMP2.PlasmaMMP2concentration is increased in obese people,andMMP2gene knockout reduces weight gain induced by a high-fat diet in mice (Bauterset al.2015).MMP13is highly expressed in adipose tissue,and its inhibitor CP-544439 can resist obesity induced by a high-fat diet in mice. Experimentsin vitroshowed that interfering with the expression ofMMP13in 3T3-L1 cells can reduce adipocyte differentiation (Shih and Ajuwon 2015). It was reported that the activity of MMPs will be specifically inhibited by the tissue inhibitors of metalloproteinase (TIMPs) (Rosen and MacDougald 2006;Guzelet al.2013). The overexpression ofTIMP3will inhibit lipid deposition in 3T3-L1 cells,accompanied by the down-regulation of some genes related to adipocyte differentiation (SREBP1C,PPARγ,etc.) (Bernotet al.2010).
Therefore,we hypothesized that TIMP2 would play an important role in lipid deposition of chickens,especially in intramuscular fat deposition of breast muscle,and it would be an important regulatory factor in the TIMPs.However,to the best of the authors’ knowledge,the physiological and molecular mechanisms of TIMPs on fat deposition in chickens have not been reported yet. In the study,we systematically explore the effect of TIMPs on IMF deposition using a co-culture system of cells from chicken muscle tissue by proteomics and high-throughput transcriptome sequencing methods.
2.Materials and methods
2.1.Animals
The Beijing-you (BJY) chickens were used,which were obtained from the Institute of Animal Sciences,Chinese Academy of Agricultural Sciences (Beijing,China). The diet was formulated according to the feeding standard(NY/T33-2004) (Appendix A). All birds were reared in the 3-story-step cages (30 birds per cage as a replicate)under the same standard conditions of lighting (20 lux),temperature (35-37°C),humidity (not less than 50%),and immunization schedule. Feed and water were providedadlibitumduring the experiment. The chickens were reared to 7 days of age for cell extraction.
2.2.Co-culture of chicken muscle satellite cells and intramuscular adipocytes
Seven chicks of 7-day-old BJY were used to extract primary cells and isolate the pectoralis major muscle for subsequent extraction. Primary cells were isolated as described previously with a slight modification (Cuiet al.2018). Cells from different animals were used for cell collection and the upper mature adipocytes were extracted from the cell suspension. The mature adipocyte layer was inoculated into a 25-cm2cell culture flask containing a complete medium. The flask was inverted for 6 days to make adipocytes adhere to the upper surface and dedifferentiate and then inverted for 6 days to make the adherent cells proliferate. After reaching 80% confluence,both cell types were sub-cultured with 0.25% trypsin EDTA (Gibco,Thermo Fisher Scientific Inc.,Suzhou,China),and the 3rd generation cells were used for further experiments.
Cells were co-cultured using Transwell plates with 0.4-μm membrane inserts (Corning Inc.,Kennebunk,ME). The two cells were initially inoculated on a separate plate,and the co-culture step was carried out according to the procedure as previously reported (Guoet al.2018).Intramuscular adipocytes (2×105/well) were inoculated in the lower hole of the 6-well plate and MSC (1×105/well)was inoculated into the upper insert to form a co-culture configuration. Cultured intramuscular adipocytes were used as the control group.
During the experiment,Eagle’s medium (DMEM)/F12(Gibco),10% fetal bovine serum (FBS,Gibco),and 1%penicillin/streptomycin (Gibco) was used as a complete medium for cell culture,and the medium was changed every 2-3 days. After 2 or 4 days of co-or single-culture,the cells were collected for follow-up experiments. All cells were incubated in a humidified atmosphere of 5%CO2at 37°C. For statistical evaluation,the 2 culture arrangements were compared 3 times.
2.3.Oil Red O staining
After 4 days of co-culture or mono-culture,the cells were washed 3 times with phosphate-buffered saline (PBS),fixed in 4% paraformaldehyde for 15 min,and then stained with a 0.5% solution of Oil Red O in 60% isopropanol for 1 h at room temperature. Afterward,the cells were washed 3 times with PBS,and the stained fat droplets in the cells were visualized by light microscopy and photographed. Stained cells were incubated in isopropanol (100%) for 10 min to extract the Oil Red O from the cells. The extracts were transferred to a 96-wellplate and the optical density at 510 nm (OD510) was measured spectrophotometrically using a SpectraMax M2 Microplate Reader (Molecular Devices,Sunnyvale,CA,USA) (Huanget al.2015).
2.4.lsobaric tags for relative and absolute quantitation (iTRAQ) assay and data analysis
Medium samples from 9 cell cultures (3 wells monoculture of intramuscular adipocytes,3 wells mono-culture MSC,and 3 wells co-culture of intramuscular adipocytes and MSC) were collected and used for iTRAQ analysis(Applied Biosystems,Foster City,CA,USA). High amounts of protein extracts were prepared,quantified by Bradford protein assay (Bio-Rad Laboratories,Hercules,CA,USA),and used for culture medium proteomic analysis after separation and detection by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).Subsequently,all samples were digested with trypsin to obtain the corresponding peptides,which were then individually labeled with the iTRAQ Reagent 8Plex One Assay Kit (AB SCIEX,Framingham,MA,USA). The liquid phase separation of the labeled peptides was performed using a Shimadzu LC-20AB Liquid Phase System(Shimadzu Corporation,Kyoto,Japan). Subsequently,peptides were desalted using a Gemini C18 solid-phase column (4.6 mm×250 mm,5 μmi.d.) and subjected to electrospray ionization tandem mass spectrometry on a Sciex TripleTOF?5600 System (AB SCIEX). Data were collected under the following conditions: spray voltage of 2 300 V,nebulizer gas at 30 psi,and spray interface temperature maintained at 150°C. Tandem mass spectrometry data were analyzed using the iQuant Software (Beijing Institute of Genomics,Shenzhen,China) and searched against the UniProt Database(Gallus gallus). A target decoy-based strategy was used to control the peptide-level false discovery rate using a threshold of 1%. Unique peptides were used for protein quantification,and normalization of protein medians was used to correct for experimental bias. The minimum number was set to 200. For protein quantification,the ratio of each sample was weighed and normalized to the control group used as the denominator for comparison.Nine cell culture medium samples were used for SDSPAGE analysis.
2.5.Analysis of candidate genes
The RNA-seq data (CRA001334) of the 14 generations of Jingxing yellow chicken IMF selection line and the RNA-seq data (CRA001908) of high-IMF and low-IMF chickens were downloaded from the BIG Data Center Database (https://bigd.big.ac.cn),and the expression of candidate genes in pectoralis muscle tissue and abdominal fat at different ages (each sample was analyzed in triplicate each age) was extracted from the CRA001334 data;after normalization of the data,the expression profile curves of candidate genes at different ages were plotted (Xinget al.2020). RNA-seq data of high-IMF and low-IMF chickens were extracted from the CRA001908 data for weighted gene co-expression network analysis (WGCNA) (Liuet al.2019).
2.6.Weighted gene co-expression network analysis (WGCNA)
The WGCNA Program Package was used to construct a weighted gene co-expression network for the gene expression values obtained from the RNA-seq of Jingxing yellow chicken pectoralis muscle. The Pearson correlation between pairs of genes was determined and a Pearson correlation matrix was constructed based on all pairs of genes (Langfelder and Horvath 2008). The optimal β-value (β=8 for the Jingxing yellow chicken) was selected to make the gene distribution conform to the scale-free network,and the weighted neighbor-joining matrix was constructed. Then,based on the correlation expression values,the adjacency relationships were converted into topological overlap matrices,and each topological overlap matrix was used for hierarchical clustering analysis.Finally,based on the topological overlap matrices,genes were clustered using the dissimilarity between genes,and the tree was cut into different gene modules (i.e.,clusters with high topological overlap) using dynamic shearing,where the smallest module should contain at least 30 genes. Co-expressed modules can be automatically color-coded by the WGCNA package,and their structure can be visualized using a heat map of topological overlap. Gene ontology (GO) term enrichment analysis of differentially expressed genes (DEGs) was performed using the DAVID functional annotation clustering method.The Kyoto encyclopedia of genes and genomes (KEGG)pathway enrichment analysis was performed using the KOBAS 3.0 online analysis tool (http://kobas.cbi.pku.edu.cn). Significance levels for GO terms and KEGG pathways were set at aP-value<0.05.
2.7.Statistical analyses
Replicates served as the experimental unit (i.e.,there were 8 replicate samples of breast muscle tissue or 3 replicate cell wells per group). The significance of differences between groups was tested by the Student’st-test using the SPSS Software version 22.0 (IBM Corp.,Armonk,NY,USA). Confidence limits were set at 95%andP<0.05 (*) orP<0.01 (**) was considered significant.All graphs were drawn using the ggplot2 Package in R 4.0.
3.Results
3.1.Lipid deposition was increased in intramuscular adipocytes in a co-culture system
In this study,we stained co-culture intramuscular adipocytes and MSC,mono-cultured intramuscular adipocytes,and mono-cultured MSC with Oil Red O to measure their lipid droplet content. The staining showed a much higher abundance of lipid droplets in intramuscular adipocytes co-cultured with MSC than in intramuscular adipocytes cultured alone,suggesting that co-culture promoted lipid deposition in intramuscular adipocytes,an effect that may have been facilitated by MSC (Fig.1-A).Lipid deposition also increased significantly in cocultured MSC (Guoet al.2018). In addition,we also analyzed the secretion of protein factors under 3 different culture conditions: MSC culture medium,intramuscular adipocytes cell culture medium,and co-culture system.Three cell culture mediums were separated by SDSPAGE,as shown in Fig.1-B. The results show that there are some highly expressed secretory proteins in the coculture system,while some proteins are not expressed in the mono-cultured intramuscular adipocytes and the mono-cultured MSC. In addition,we also measured the protein expression of TIMP2 in mono-cultured MSC culture medium,mono-cultured intramuscular adipocytes culture medium,and co-culture cell culture medium. The results showed that TIMP2 was expressed in monocultured MSC culture medium and co-culture cell culture medium,and there was almost no expression in monocultured intramuscular adipocytes culture medium(Fig.1-C).
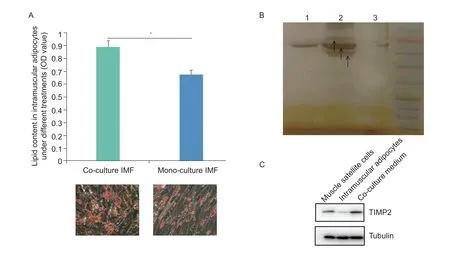
Fig.1 The results of Oil Red O and protein expression through co-culture system. A,Oil Red O staining showed that co-culture Intramuscular fat (IMF) cells contained more lipid droplets on day 4. The OD510 values of Oil Red O uptake by co-culture and monoculture IMF cells on day 4 (n=3). The data are expressed as the mean±SD (*,P<0.05). B,the results of SDS-PAGE analysis.Isolation of serum-free cell cultures showed that more (types and amounts) secreted proteins were present in myoblast cultures after 2 days of culture alone than in the co-culture system. Lane 1,the culture medium of mono-cultured MSC;lane 2,the cocultured cell culture medium;lane 3,the culture medium of mono-cultured intramuscular adipocytes. C,results of Western blot in muscle satellite cells,intramuscular adipocytes,and co-culture medium.
3.2.Tissue inhibitor of metalloproteinases 2(TlMP2) protein level was increased in the co-culture system
We also determined the number of proteins secreted into the culture medium by mono-cultured intramuscular adipocytes and MSC,as well as by intramuscular adipocytes co-cultured with MSC. The results showed that 54 secreted proteins were detected in the monocultured intramuscular adipocyte medium,93 secreted proteins were detected in the mono-cultured MSC medium,and 25 secreted proteins were detected in the co-cultured cell system medium. The results of a comparative analysis revealed that 6 proteins were only present in the intramuscular adipocyte mono-culture medium,namely TKT,CMPK,RCN1,CALD1,LTBP1,and YWHAQ. There were 43 proteins only present in MSC mono-culture medium,including ACLY,GAPDH,GOT2,HSP90B1,MMP2,PSMA4,YWHAE,COMP,VCP,and etc. TIMP2 protein was only detected in the co-culture system medium (Fig.2-A;Appendix B).
After differential expression analysis and identification,54 differentially expressed proteins were identified in the isolated intramuscular adipocyte culture medium and co-culture system cell culture medium. After co-culture,the expression levels of TIMP2,PKM,THBS2,and other proteins increased,while the expression levels of APOA1,BASP1,COL12A1,COL5A1,COL6A2,and other proteins decreased. 84 differentially expressed proteins were identified in MSC mono-culture medium and coculture system medium. Meanwhile,the KEGG pathway enrichment analysis of differentially expressed proteins in the two groups showed that they were simultaneously enriched in ECM receptor interaction,focal adhesion,glycolysis,and other signaling pathways (Fig.2-B and C;Appendix C).
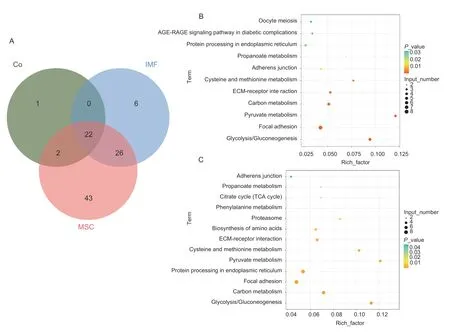
Fig.2 Protein Venn diagram and KEGG pathway analysis of mono-cultured intramuscular adipocyte medium,mono-cultured muscle satellite cells (MSC) medium,and co-culture system medium. A,protein Venn diagrams between mono-cultured intramuscular adipocyte medium (IMF),mono-cultured MSC medium (MSC),and co-culture system medium (Co). B,the pathways of monocultured intramuscular adipocyte medium and co-culture system medium were enriched differently. C,the pathways of monocultured MSC medium and co-culture system medium were enriched differently.
3.3.TIMP2 gene expressed in pectoralis muscle tissue
The developmental expression profile of theTIMP2gene was also analyzed using RNA-seq data of pectoralis muscle tissue and abdominal fat tissue of Jingxing yellow chickens at different ages. The results showed that the expression of theTIMP2gene peaked at 98 days in pectoralis muscle tissue. Moreover,the expression ofTIMP2was significantly higher (P<0.05) in abdominal fat tissue than in pectoralis muscle tissue (Fig.3-A;Appendix D). In addition,the transcriptome sequencing analysis of pectoralis muscle tissues with high-and low-IMF content revealed that the expression of theTIMP2gene in pectoralis muscle tissues with high-IMF content was also significantly higher (P<0.05) than that in pectoralis muscle tissues with low-IMF content (Fig.3-B).
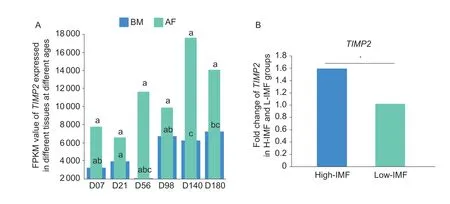
Fig.3 The expression of TIMP2 in Jingxing yellow chickens. A,the developmental expression profile of TIMP2 in breast muscle(BM) tissue and abdominal fat (AF) of Jingxing yellow chickens (n=3). Peer data with different lowercase letters on shoulder labels indicate significant differences (P<0.05). B,the expression of TIMP2 in the RNA-seq data of pectoralis muscle tissue with high-Intramuscular fat (H-IMF) and low-Intramuscular fat (L-IMF) content of Jingxing yellow chickens (n=8). The data are expressed as the fold change (*,P<0.05).
3.4.TIMP2 gene expression affects the process of lipid deposition
To further examine the correlation between expression of genes and phenotypes,we performed a WGCNA of all genes expressed in the high-and low-IMF pectoralis muscle tissue transcriptome data. During the analysis,we constructed a co-expression network using raw values of the fragments per kilobase of feature per million mapped reads (FPKM) from a total of 16 individuals,ultimately identifying 23 modules (Fig.4;Appendix E).According to the results,we first selected the module where the candidateTIMP2gene was located,in which turned out to be the red module. As shown in Fig.4,the red module was highly correlated with IMF trait (R=0.84;P=5e-05),TG level (R=0.83;P=6e-05),fatty acid C14:0 content (R=0.77;P=5e-04),C14:1 content (R=0.73;P=0.001),C16:0 content (R=0.62;P=0.01),C16:1 content (R=0.85;P=4e-05),C18:1n9c content (R=0.86;P=2e-05).
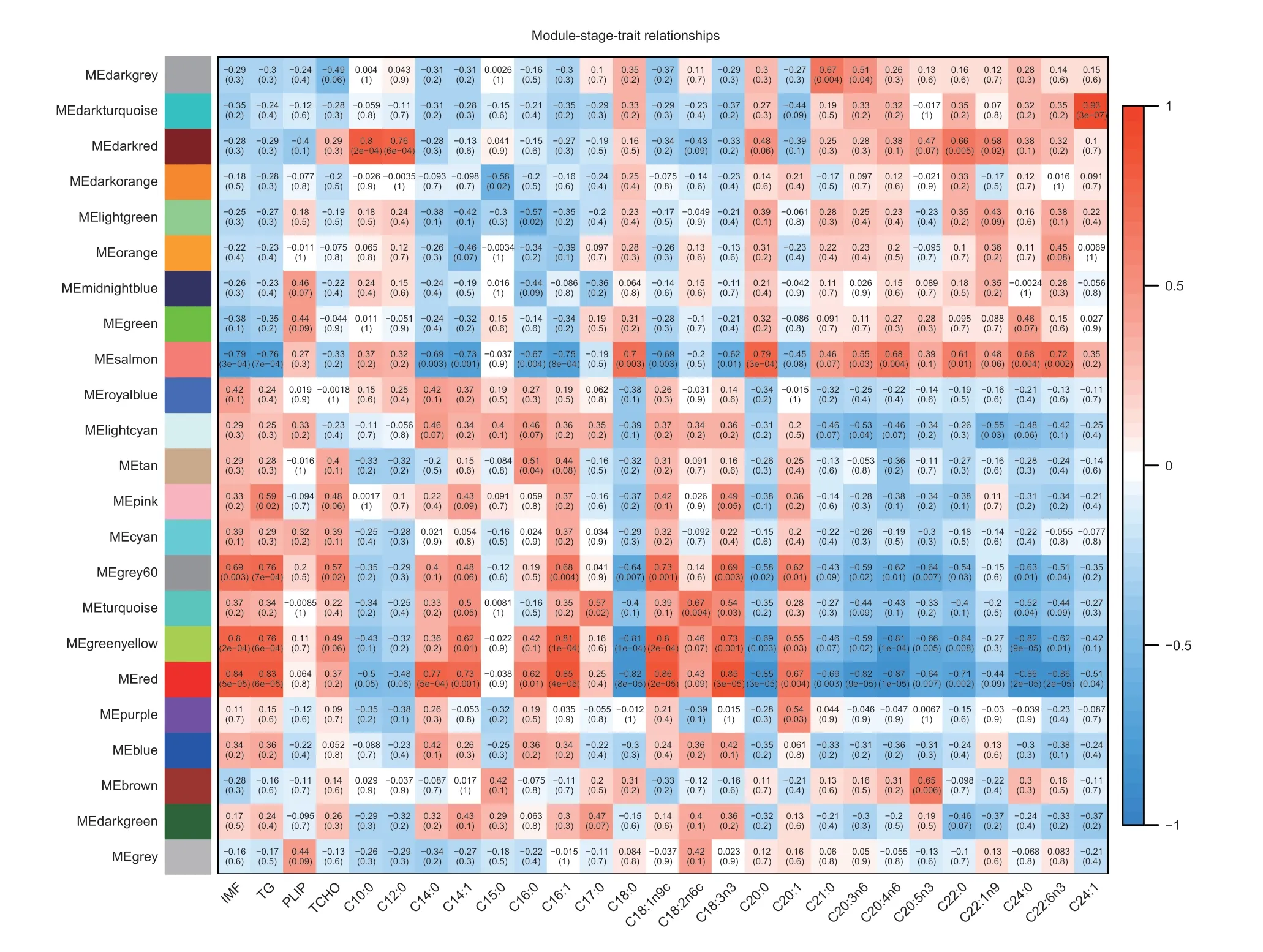
Fig.4 Module-stage-tissue relationship of high-Intramuscular fat (H-IMF) content and low-Intramuscular fat (L-IMF) content. Each cell contains the corresponding correlation and P-value. The table is color-coded by correlation according to the color legend. TG,triglyceride;PLIP,phospholipids;TCHO,total cholesterol;ME,module eigengenes.
3.5.TIMP2 gene was co-expressed with lipid deposition-related genes
In addition,the analysis of the other genes in the red module revealed thatTIMP2was co-expressed with some lipid metabolism-related genes,such asADIPOQ,SCD,ELOVL5,ELOVL7,andLPL,and collagenase-related genes,such asCOL1A2,COL4A2,COL5A1,COL6A1,andCOL6A3. These findings suggest that theTIMP2gene has similar expression patterns as those of these genes and may have similar regulatory effects. These genes were also subjected to differential expression analysis and found to be significantly upregulated (P<0.05)in high-IMF pectoralis muscle tissue. These results further indicate thatTIMP2promotes lipid deposition(Fig.5;Appendices F and G).

Fig.5 Chord diagram of the correlation between the co-expression of TIMP2 and lipid metabolism-related genes in the red module.
3.6.Pathway enrichment analysis of TIMP2 gene and its co-expression genes
To further understand the relationships betweenTIMP2and its co-expressed genes,we conducted further functional enrichment analysis of all genes in the red module whereTIMP2resides. Significant GO conditions and pathways (P<0.05) are listed in Appendix H. As it can be seen,these co-expressed genes are significantly enriched in GO terms and ECM-related pathways.Among these GO terms,13 are related to the ECM,such as ECM tissue,extracellular space,etc.(Fig.6-A). KEGG pathway enrichment analysis showed that there were 16 pathways enriched in red module genes (P<0.05),which comprised many pathways related to cell connection and membrane structure,such as adhesion junction,vascular smooth muscle contraction,gap junction,ECM receptor interaction,and cell adhesion molecules (CAMs). These findings suggest that proteins encoded by the genes in this module may interact with ECM and other signaling pathways (Fig.6-B;Appendix I).
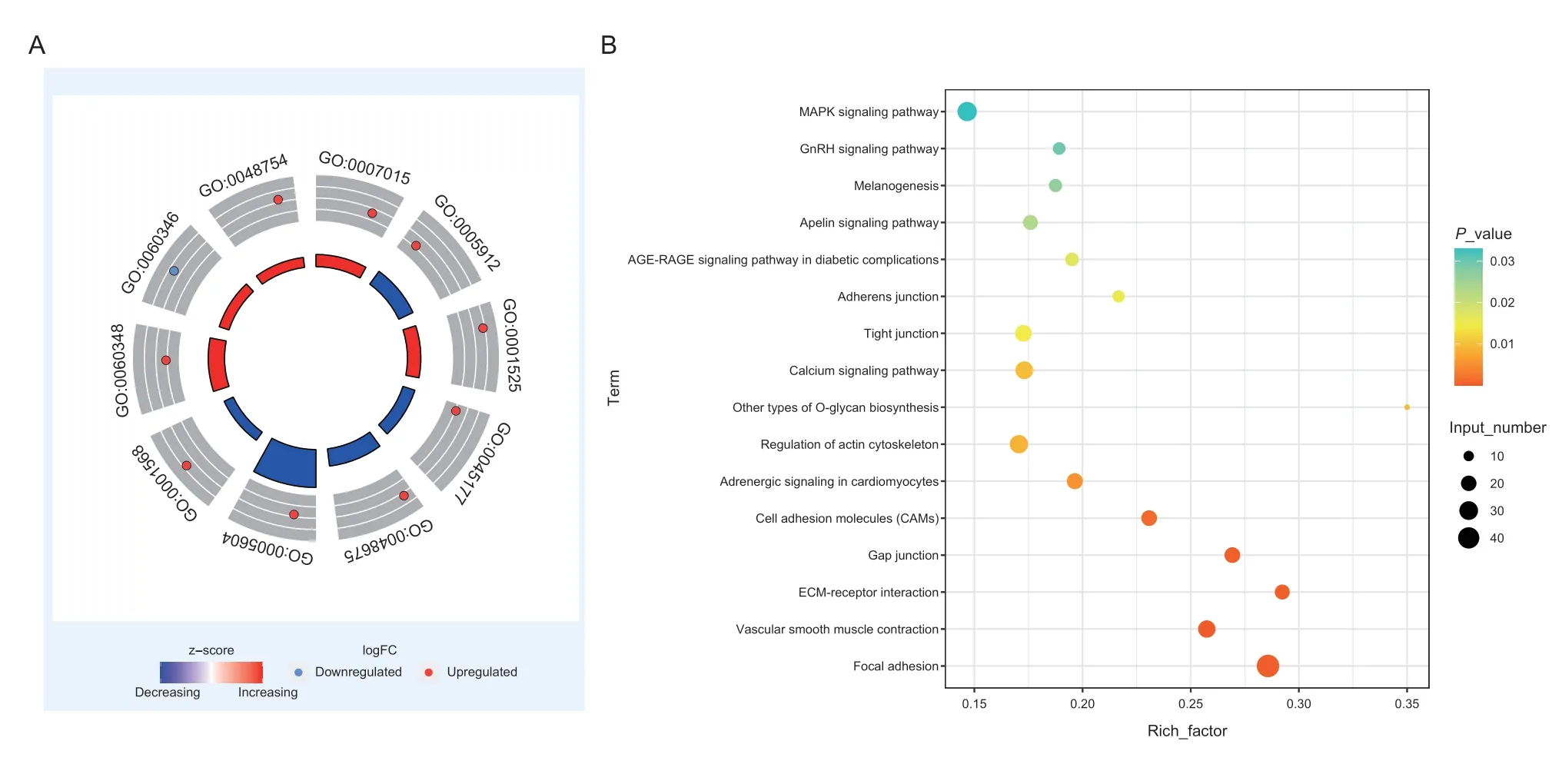
Fig.6 GO enrichment circle plots and KEGG pathway enrichment bubble plots for the red module. A,GO enrichment circle plots for the red module. The inner circle shows the z score,the outer circle shows the GO ID,inside the outer circle is the upregulation and downregulation distribution of participating genes. B,KEGG pathway enrichment bubble plots for the red module. The x-axis represents the enrichment factor (Enrichment factor=Number of genes enriched in the pathway/Number of all genes in the background genome),the y-axis represents the enriched pathway. The colors represent the enrichment significance and the size of the bubble represents the number of genes enriched in the pathway.
4.Discussion
The results of the present study have supported our hypotheses that TIMP2 would play an important role in lipid deposition of chickens,especially in intramuscular fat deposition of breast muscle,and it would be an important regulatory factor in the TIMPs. Through proteome sequencing,we determined that TIMP2 is an important regulatory factor in the TIMPs family.Meanwhile,with the increase of TIMP2 protein content,the lipid droplets in the co-cultured intramuscular adipocytes increased. So we can preliminarily confirm that TIMP2 promoted the deposition of intramuscular adipocytes. Transcriptome sequencing results of breast muscle tissue at different ages showed thatTIMP2gene content in abdominal fat tissue was significantly higher than that in breast muscle tissue,further confirming thatTIMP2has a regulatory effect on fat deposition. WGCNA was conducted on the transcriptome sequencing results of breast muscle tissue with high-and low-IMF. It was found that the module whereTIMP2was located was significantly positively correlated with the IMF content of breast muscle tissue. In this module,TIMP2was coexpressed with some genes related to lipid metabolism.These results provide a scientific basis for the regulation ofTIMP2on intramuscular fat deposition in breast muscle tissue.
In general,the key candidate proteins TIMP2 affecting IMF deposition were screened by quantitative proteomic analysis for the first time. The candidate genes were verified by analyzing the expression profiles of pectoralis muscle tissues of Jingxing yellow chickens at different ages and the results of the sequencing analysis of pectoralis muscle tissues with high-and low-IMF content. WGCNA and enrichment analysis was performed to comprehensively examine the interaction mechanism between intramuscular adipocytes and MSC. The results of Oil Red O staining showed that the content of lipid droplets in co-cultured intramuscular adipocytes was significantly higher than that in single cultured intramuscular adipocytes. This suggests that MSC may promote lipid deposition of intramuscular adipocytes. According to the published researches,interfering with the structure of the cell membrane can significantly affect the differentiation of adipocytes.Matrix metalloproteinases (MMPs) are important enzymes for degradation of ECM (Evrosimovskaet al.2012;Chienet al.2013). Among them,MMP2is one of the family members with outstanding degradation of ECM. At the same time,the activity of MMPs was inhibited by tissue inhibitors of metalloproteinase(TIMPs) (Visse and Nagase 2003;Guzelet al.2013).ThusTIMP2can interfere with the structure of the cell membrane by acting onMMPsand thus affect lipid deposition. In this study,TIMP2was found to be differentially expressed in pectoralis muscle tissues with high-and low-IMF content,as well as in pectoralis muscle tissues at different ages. Changes or upregulation in TIMP2 expression can lead to the accumulation of ECM proteins,ultimately resulting in tissue fibrosis (Yasmeenet al.2019).
In addition,developmental expression profiles of the pectoralis muscle and abdominal adipose tissue showed that the expression of theTIMP2gene in abdominal adipose tissue was significantly higher than that in breast tissue,and the expression trend in the two tissues was the opposite. The WGCNA also identified genes associated with lipid deposition-related phenotypes and revealed that theTIMP2gene was highly positively correlated with the IMF content,TG level,and C14:0 content phenotypes. Fatty acid synthesis and deposition were also significantly increased,suggesting thatTIMP2may be involved in the regulation of lipid metabolism in chicken muscle. Furthermore,TIMP2is co-expressed with classic genes related to fat metabolism in the red module,which is involved in the biological processes of fatty acid metabolism (CPT2,ELOVL5,ELOVL7,and etc.). This finding suggests that theTIMP2gene shares similar expression patterns with these genes and therefore may play a similar regulatory role together. Through the enrichment and analysis of differentially expressed genes in the red module,we identified signaling pathways related to lipid metabolism,including ECM receptor interaction signaling pathway and various pathways related to cell connection and membrane structure,indicating that the genes in the red module may play a role through these signaling pathways. ECM receptor interactions also have profound implications for major cellular processes,including cell growth,differentiation,migration,and survival (Huanget al.2021). As the support of cell structure,ECM can affect cell morphology and regulate cell metabolisms,such as migration,proliferation,differentiation,and information transmission(Zhanget al.2009). A study by Leeet al.(2013) found that fibronectin and integrin from different members of the ECM were upregulated in different adipose tissues,and that collagen and whole con-junction were significantly enriched in subcutaneous and intramuscular adipose tissues,respectively. However,due to the difficulty of the analysis of the ECM composition,not much attention has been paid to fat composition to date. Thus,the theory that the expression level of theTIMP2gene affects lipid deposition through cell membrane structure was further confirmed.
Based on a co-culture system of intramuscular adipocytes and muscle satellite cells,we simulated the physiological system between skeletal muscle and adiposity and explored their interactions. The results showed that TIMP2,an important protein screened,has a facilitative effect on lipid deposition in muscle adipocytes.This experiment combines proteome sequencing analysis and transcriptome data analysis,which is helpful to deeply understand the interaction between muscle and fat.Afterward,more research will be carried out to provide important insights into the research of livestock meat quality.
5.Conclusion
TIMP2,as a key inhibitor of matrix metalloproteinases,regulates IMF deposition in a chicken muscle through the ECM-receptor interaction pathway under the influence of muscle satellite cells. The results of the study provide a new research idea for the molecular mechanism of chicken lipid deposition,and the interaction between intramuscular adipocytes and muscle satellite cells,which will provide a theoretical basis for the high-quality production of poultry meat in the future.
Acknowledgements
This research was funded by the grants from the National Natural Science Foundation of China(31872340),the State Key Laboratory of Animal Nutrition(2004DA125184G2109),the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (ASTIP-IAS04),and the earmarked fund for China Agriculture Research System (CARS-41).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was conducted by the Guidelines for Experimental Animals Established by the Ministry of Science and Technology (Beijing,China). All experimental protocols were approved by the Science Research Department (in charge of animal welfare issues) of the Institute of Animal Sciences,Chinese Academy of Agricultural Sciences (CAAS,Beijing,China) (No.IAS2019-21).
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
 Journal of Integrative Agriculture2023年3期
Journal of Integrative Agriculture2023年3期
- Journal of Integrative Agriculture的其它文章
- Germinated brown rice relieves hyperlipidemia by alleviating gut microbiota dysbiosis
- Melatonin treatment alleviates chilling injury in mango fruit 'Keitt'by modulating proline metabolism under chilling stress
- Changes in the activities of key enzymes and the abundance of functional genes involved in nitrogen transformation in rice rhizosphere soil under different aerated conditions
- Growth and nitrogen productivity of drip-irrigated winter wheat under different nitrogen fertigation strategies in the North China Plain
- Effect of fertigation frequency on soil nitrogen distribution and tomato yield under alternate partial root-zone drip irrigation
- Phylogenetic and epidemiological characteristics of H9N2 avian influenza viruses in Shandong Province,China from 2019 to 2021
