StOFP20 regulates tuber shape and interacts with TONNEAU1 Recruiting Motif proteins in potato
Al Ju ,WANG Ye ,YAN Ya-wen,Ll Chen-xiao,LUO Wei,MA Ling,SHANG Yi,GAO Dong-li
Yunnan Key Laboratory of Potato Biology,the CAAS-YNNU-YINMORE Joint Academy of Potato Science,Yunnan Normal University,Kunming 650500,P.R.China
Abstract The OVATE family proteins (OFPs) are plant-specific proteins that modulate diverse aspects of plant growth and development. In tomato,OFP20 has been shown to interact with TONNEAU1 Recruiting Motif (TRM) proteins to regulate fruit shape. In this study,we demonstrated that the mutation of StOFP20 caused a shift from round to oval shaped tubers in a diploid accession C151,supporting the role of StOFP20 in controlling tuber shape. Its expression reached a maximum in the tuber initiation stage and then decreased as the tuber develops. To help elucidate the mechanism of tuber shape regulation by StOFP20,27 TONNEAU1 Recruiting Motif (TRM) proteins were identified and 23 of them were successfully amplified in C151. A yeast two-hybrid assay identified three TRM proteins that interacted with StOFP20,which was confirmed by firefly luciferase complementation in tobacco leaves. The OVATE domain was indispensable for the interactions,while the necessity of the M10 motif in TRM proteins varied among the interactions between StOFP20 and the three TRMs. In summary,both StOFP20 and SlOFP20 directed interactions with TRM proteins,but the corresponding interactants were not completely consistent,implying that they exert regulatory roles through mechanisms that are only partially overlapping.
Keywords: potato,tuber shape,OFP20,TRM
1.lntroduction
Potato tuber shape constitutes an important component of tuber quality for both fresh market use and the processing industry. Cultivated potato displays a rich diversity in shape,and eight basic categories of compressed,round,ovoid,obovoid,elliptic,oblong,long-oblong and elongated,have been documented by the International Potato Center (CIP). The phenotypic variability in tuber shape allows genetic analysis of this important trait,andRowas found to be a major gene conferring round tuber shape. van Ecket al.(1994) initially reported the identification ofRoon chromosome 10,and noted that the round shape was dominant over the long shape. A SNP marker highly associated with theRolocus was identified,and 375 genes surrounding thelocus were annotated (Lindqvist-Kreuzeet al.2015). Wuet al.(2018) demonstrated thatSlOFP20underlies fruit shape in tomato,and further showed thatRowas fine-mapped to a syntenic region containingSlOFP20andStOFP20markers that were genetically associated with tuber shape. However,transgenic confirmation of the function ofStOFP20has not been established.
OFP20 is a member of the OVATE family protein class. Prior to the identification ofSlOFP20,OVATEwas cloned in tomato and has been shown to control oval shape (Liuet al.2002). TheOVATEgene encodes a hydrophilic protein with a C-terminal domain of about 70 amino acids which is designated as the OVATE domain.Both SlOFP20and OVATE interact with TONNEAU1 Recruiting Motif (TRM) proteins (Wuet al.2018). TRM,TONNEAU1 (TON1) and a Protein Phosphatase2A (PP2A)holoenzyme containing FASS/TONNEAU2 (TON2) as a regulatory subunit constitute a TTP complex. This complex is required for the formation of a specialized microtubule apparatus-preprophase band (PPB) that organizes the cell division plane (Spinneret al.2013).Disruption ofTON1orFASS/TON2impairs PPBs in premitotic cells,conferring dwarf and misshapen phenotypes inArabidopsismutants (Camilleriet al.2002;Azimzadehet al.2008). In addition to the absence of PPBs,TON2mutation also affects cortical microtubule reorientation in interphase cells (Kiriket al.2012). TRM proteins are generally responsible for the microtubule recruitment of TTP protein components (Drevenseket al.2012). TheArabidopsisgenome encodes 34 TRM proteins,but not all of them can bind microtubules which reinforces their diversified functions (Kiriket al.2012).LONGIFOLIA1 (TRM1) and LONGIFOLIA2 (TRM2)regulate the shapes of aerial parts inArabidopsis(Leeet al.2006). In rice,GW7 encodes a TRM protein,and the upregulation ofGW7results in the production of more slender grains and enhanced rice grain appearance quality (Wanget al.2015). Given the importance of TRMs in determining organ shape,an analysis of TRMs in potato will help to elucidate the mechanism of tuber shape regulation.
Previously,we developed a diploid potato population segregating for tuber shape. Map-based cloning delimited the causative genes to an interval containingStOFP20,and PCR amplification revealed thatStOFP20was absent in the oval tubers but present in the round tubers (Aiet al.2020). In this study,we provided evidence thatStOFP20controls tuber shape,and analyzed the expression ofStOFP20over the course of young tuber development.To shed light on the mechanism of StOFP20 regulating tuber shape,TRM proteins interacting with StOFP20 were identified and interactions between TRM and TON proteins were further investigated.
2.Materials and methods
2.1.Generation of StOFP20-edited transgenic plants
The two 20-ntguide RNA sequences were 5′-AAAAGACAACCCCATAACCT-3′ and 5′-AAAGGCGTA GTACCAATTCC-3′. The target sequence was introduced into the CRISPR/Cas9 vector pKSE401 carrying theArabidopsisU6 promoter. C151 (CIP705468) belongs toSolanum tuberosumand was provided by CIP in Lima,Peru. Preculture medium containing 4.43 g L-1MS,20 g L-1sucrose,1 mg L-1zeatin riboside (ZR) and 2 mg L-1α-naphthaleneacetic acid (NAA) was prepared in Petri dishes,two sterilized papers were placed on the medium and 2 mL PACM solution containing 0.44 g L-1MS,0.2 g L-1caseine hydrolysate,3 g L-1sucrose,100 μL 2,4-D (1 mg mL-1) and 50 μL kinetine(1 mg mL-1) was supplemented onto the papers. The stems of 4-wk-old C151 plants were placed on the moist sterilized papers for 2 d. During this period,cultures for agrobacterium strain GV3101 carrying the binary plasmid were prepared. Then the explants were immersed in the agrobacterium culture with OD600=0.45-0.6. After the addition of 40 mg L-1AS for 10 min,the explants were kept dry on the filter paper before being transferred to the preculture medium with the addition of 40 mg mL-1AS in the dark for 2 d. After 2 d,the explants were placed on the cultivation medium containing 4.43 g L-1MS,20 g L-1sucrose,2 mg L-1ZR,0.01 mg L-1NAA,200 mg L-1timentin and 100 mg L-1kanamycin. After about 4 wk,the emerging shoots were transferred to the rooting medium that contained 4.43 g L-1MS,30 g L-1sucrose,200 mg L-1timentin,and 50 mg L-1kanamycin.
DNA was extracted from the rooted plants and the presence of Cas9 was examined. StOFP20 was amplified from plants harboring the Cas9 protein and the PCR products were TOPO-cloned to examine the mutation types ofStOFP20. The mutants were grown in a greenhouse located in the university. Expanded leaves collected from the mutants and control plants were subject to ploidy examination by flow cytometry (Alsahlanyet al.2019). Tubers were harvested when the plants reached maturity and their lengths and widths were measured.Length was measured from the stolon initiation to the end of the tuber,and width was measured perpendicular to the length. Five tubers were measured for each plant.
2.2.Gene expression analysis by qRT-PCR
Samples were harvested from plants homozygous for the round tuber. RNA was extracted using the RNeasy Plant Mini Kit and treated with DNaseI (Qiagen,China). Firststrand cDNA synthesis (TaKaRa,China) was performed with 1.5 μg of total RNA for each reaction. qPCR was performed using TB Green Premix (TaKaRa) to examine the expression ofStOFP20. The relative expression levels were calculated using the comparative Ct (2-ΔΔCt)method (Schmittgenet al.2008).
2.3.Phylogenetic and protein motif analyses of TRMs
To retrieve all putative TRM proteins in potato,the protein sequences of 34 members of this family inArabidopsiswere blasted against the Potato PGSC DM v3.4 protein sequences (http://solgenomics.net/) at a cutoff E-value of 10-10. After removing redundant protein sequences,the representative transcripts were searched for the remaining 49 proteins in the Spud DB Potato Genomics Resource (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml). Multiple alignments of the full-length amino acid sequences of 22 TRMs fromArabidopsisand potato were performed using Clustalw2 (https://www.ebi.ac.uk/Tools/msa/clustalw2/) with the default parameters.PGSC0003DMP400054403 was excluded due to its short length. The phylogenetic relationships among all the TRMs were estimated using MEGA X (Kumaret al.2018)with the neighbor-joining method based on the p-distance and bootstrap tests replicated 1 000 times. The same method was used to analyze the OFP proteins. The MEME tool (Baileyet al.1994) was used to discover the conserved motifs with the following parameters: “nmmotifs 8,minw 10,maxw 100,minsites 30,maxsites 120”.
2.4.Yeast two-hybrid (Y2H) analyses
Full-lengthStOFP20was cloned into the pGBKT7 vector by T4 DNA ligase as a C-terminal fusion to the GAL4 DNA binding domain (BD). Full-lengthTRMswere cloned into the pGADT7 vector by in-fusion cloning as a C-terminal fusion to the GAL4 DNA activation domain (AD). TheStOFP20coding sequences ranging from 1-849 bp(StOFP20-M1) and from 850-1 020 bp (StOFP20-M2)were separately integrated into the pGBKT7 vector by infusion cloning. The coding sequences for each of the three TRMs were dissected into two fragments,and each fragment was obtained by PCR amplification. Then infusion cloning was used to integrate the two fragments lacking the M10 domain into the pGADT7 vector. The primers for the cloning of the 23 StTRMs and other primers are listed in Appendix A. The constructs were then transformed into the Y2HGold yeast strain which was plated on SD/-Trp-Leu medium and SD/-Trp-Leu-His medium. Colonies able to grow on SD/-Leu-Trp-His (SD/-LTH) medium were inoculated in the YPDA medium with the addition of aureobasidin A (AbA) for 24 h.Bacterial cultures of 2 μL were plated on SD/-Trp-Leu-His+AbA medium.
2.5.Transient expression of proteins in Nicotiana benthamiana
The full-length coding sequence ofStOFP20and the three interactingTRMcDNA were introduced into the pCAMBIAnLuc and pCAMBIA-cLuc vectors,respectively. The binary plasmids were transformed into theAgrobacterium tumefaciensstrain GV3101. After 2 d of co-infiltration,the luciferase substrate D-luciferin (L9504,Sigma,USA) was sprayed onto the leaf surface. The chemiluminescence signal was observed using Tanon5200 Multi after 5 min of incubation in the dark.
3.Results
3.1.Loss of function of StOFP20 alters tuber shape
StOFP20is an intronless gene and its coding sequence comprises 1 041 bp. The similarities between StOFP20 and SlOFP20 were 83% in coding sequence and 80%in protein sequence (Appendix B). To confirm the biological function ofStOFP20,loss-of-function mutants were generated using CRISPR/Cas9 technology (Xinget al.2012). A construct expressing two guide RNAs targeting the 166-188 bp ofStOFP20wasintroduced into the diploid accession C151. Four independent transformants were generated and their PCR products were sequenced (Fig.1-A). Mutants with a single peak in the sequencing chromatogram were considered homozygous,while mutants with double peaks in the sequencing chromatogram were considered to be either heterozygous with both wild type and mutated alleles or mosaic with more alleles than expected. Sequencing of theofp20-1PCR products revealed a single peak in the sequencing chromatogram for the first target,indicative of the homozygous mutation (Appendix C),and the 1-bp insertion resulted in a premature stop codon (Appendix D). For the remaining mutants,double peaks were observed for the first target (Appendix C),hence multiple bacterial clones were sequenced to further explore the variability of the edition profiles. Forofp20-2,seven colonies were sequenced;one allele was the same as the wild type,four alleles contained a premature stop codon,and two contained frame-shift mutations. Forofp20-3,seven colonies were sequenced;two alleles harbored a premature stop codon,and five alleles harbored frame-shift mutations. Forofp20-4,nine colonies were sequenced: Seven alleles showed a premature stop codon and two alleles showed frameshift mutations (Appendix D). Potato transformation through callus formation tends to change the ploidy level of plantlets. Ploidy detection by flow cytometry showed that the four mutants became tetraploids (Appendix E). Because OFP20was completely absent in the oval tubers of the C151 population,in theory at most two alleles should be detected in the mutants. The presence of multiple alleles in the mutants indicated chimerism between the different edited cell-lines in the analyzed tissues.ofp20-5contained the sequence ofCas9but the targeted sequence was not mutated,hence it was selected as a transgenic tetraploid control (Appendix C).Visual inspection found that the four mutants bear oval tubers,whereas the control set round tubers (Fig.1-B).Measurements of the tuber length/width ratio showed that the average length/width ratio was 0.84 for the untransformed diploid control and 0.95 for the unmutated tetraploid control,while the ratios ranged from 1.29 to 1.72 for the transgenic plants (Fig.1-C). The transgenic evidence confirmed thatStOFP20controls tuber shape.
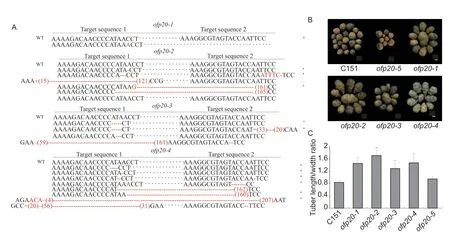
Fig.1 CRISPER/Cas9-edited StOFP20 alters the tuber shape of a diploid accession C151. A,edition profiles of StOFP20 in the transgenic plants. The type of mutation is denoted by a red letter for small insertion,+for large insertion where the number inside the bracket means the number of inserted nucleotides,-for deletion where the number of short dashes means the number of deleted nucleotides for small deletion,and the number inside the bracket means the number of deleted nucleotides for large deletion. * indicates mutated alleles containing a premature stop codon. Some other colonies showed the same variations as listed,so their sequences are not displayed. For ofp20-1,a homozygous mutation was detected in the first target,and the 1-bp insertion resulted in an early stop codon,so sequencing of the second target was not performed. B,visual inspection of tubers harvested from transgenic and control plants. Bar=1 cm. C,length/width ratio measurements of mature tubers harvested from mutants and controls. Error bars represent the standard deviations of five tubers for each plant. * and **,P<0.05 and P<0.01,respectively.
3.2.Expression pattern of StOFP20
Tuber shape is determined from an early time point in tuber development (Xuet al.1998). To understand the role ofStOFP20at the early stage of tuber development,samples were collected over five stages from stolons and young tubers,generally in accordance with the stages from stage 2 to 6 as described by Kloostermanet al.(2005). qRT-PCR was carried out to determine the transcript abundance ofStOFP20,revealing that itsexpression increased with swelling of the stolon,peaked at the tuber initiation stage and then decreased as the young tuber develops. In addition,qRT-PCR was performed in various organs to determine the broad role ofStOFP20in organ development. It was found thatOFP20RNA accumulated predominantly in flowers and to a lesser extent in roots and only marginally in stems and leaves (Fig.2).
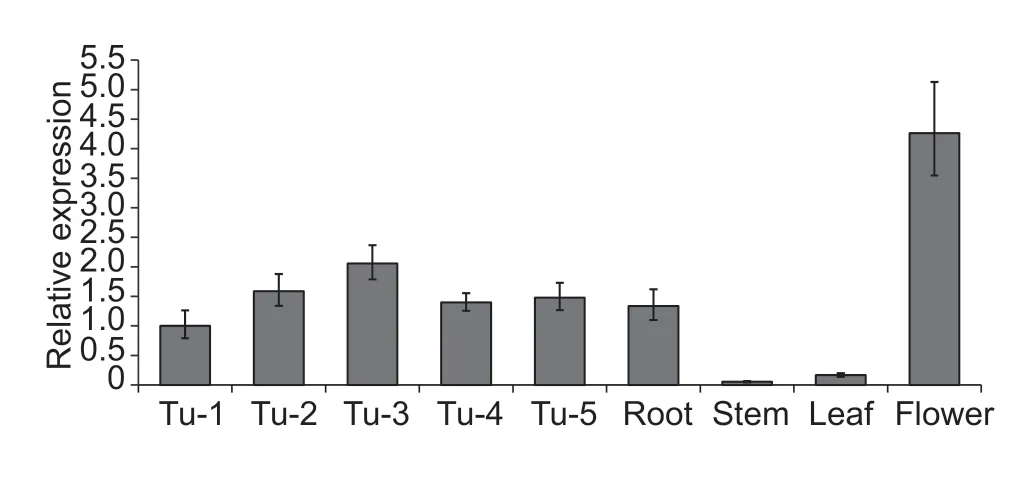
Fig.2 Expression of StOFP20 in developmental stages and organs. Tu-1 to Tu-5 represent tuber developmental stages. Tu,tuber;Tu-1,no swelling in the subapical region of the stolon;Tu-2,slight subapical swelling;Tu-3,prominent subapical swelling(tuber initiation stage);Tu-4,enlarged tubers with a diameter of~0.5 cm;Tu-5,enlarged tubers with a diameter of~0.8 cm.The potato Actin gene was used as an internal control. Error bars represent standard deviations of three biological replicates.
3.3.ldentification of TRM proteins interacting with StOFP20
To explore the possibly of conserved molecular mechanisms of SlOFP20 and StOFP20 in regulating harvestable organ shape,we performed a genome-wide identification of TRMs in the DM genome. A total of 27 TRM proteins were identified using theArabidopsisTRM protein sequences for the Blastp search,and ultimately 23 of them were successfully amplified in C151. The phylogenetic relationships of the TRM proteins inArabidopsisand potato are presented in Fig.3-B. With an aim of searching for the motifs in the 23 TRM proteins,the parameter set described by Wuet al.(2018) was used. An M10 motif with a protein sequence similar to the M8 motif present in tomato TRM proteins was detected(Fig.3-A). Comparison of the abundance of the M10/M8 motif showed that 11 potato TRM proteins contain the M10 motif (Fig.3-B) and 14 tomato TRM proteins harbor the M8 motif (Wuet al.2018).
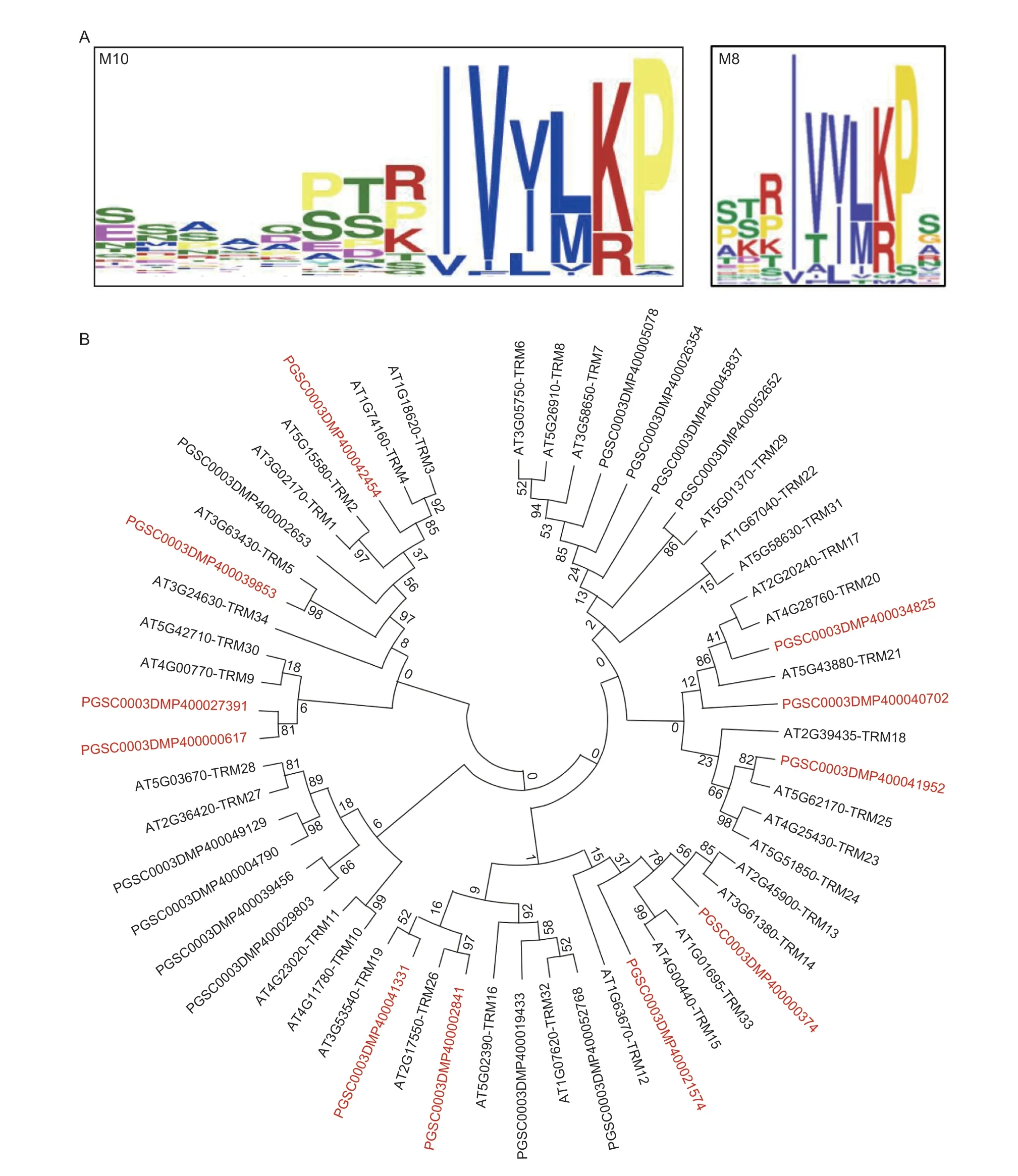
Fig.3 Analysis of TRM proteins in potato. A,sequence of the M10 motif in potato TRM proteins (left panel) and sequence of the M8 motif in tomato TRM proteins (right panel). B,phylogenetic analysis of TRM proteins in Arabidopsis and potato. Proteins prefixed with AT were identified from Arabidopsis,and PGSC were from potato. TRM proteins containing the M10 motif are shown in red.
To identify the StOFP20 protein partners,a pairwise Y2H assay was performed with the 23 successfully amplified TRMs. Since the interaction of SlOFP20 and SlTRM5 was reported,they were included as a positive control. StOFP20 interacted with StTRM5(PGSC0003DMP400039853),StTRM19 (PGSC0003 DMP400041331),and StTRM20 (PGSC0003DMP400034825)when yeast was grown on SD/-LTH medium with 125 ng mL-1AbA (Fig.4-A). To confirm this interaction,StOFP20 and three TRM proteins were fused separately with the N-terminal half(OFP20-nLuc) and the C-terminal half (cLuc-TRM) of firefly luciferase. TRM11 (PGSC0003DMP400029803),which did not interact with StOFP20 in the Y2H assay,was employed in the negative control. As shown in Fig.4-B,luciferase activity could be detected after co-expression of StOFP20-nLuc and cLuc-StTRMs,indicating the direct interactions of StOFP20 and the three TRM proteinsin vivo(Fig.4-B). No interactions were detected between StOFP20 and the other 20 TRMs in yeast.
To test whether the OVATE domain is necessary for the interactions between OFP20 and the TRM proteins,truncated versions of StOFP20 were cloned into twohybrid vectors. In the OFP20-M1 vector,the 850-1 041 bp C-terminal fragment harboring the OVATE domain was deleted,and in the OFP20-M2 vector,only the 850-1 020 bp OVATE domain was integrated (Fig.5-A). The Y2H assay revealed that the conserved OVATE domain was necessary and sufficient for interactions with the three TRM proteins (Fig.5-B). The necessity of the M10 motif was investigated by a deletion of the 14 aa M10 motif in the three TRM proteins (Appendix F). It was found that the absence of the M10 motif in the TRM19 and TRM20 proteins did not affect their interactions with StOFP20,but in contrast,deletion of the M10 motif abolished the interaction between StOFP20 and TRM5 (Fig.5-B).
3.4.A putative TON2 interacts with TRM proteins
TRM1 is able to target TON1 and TON2 to cortical microtubulesviaits C-terminal TON interaction motif(Drevenseket al.2012;Spinneret al.2013). To analyze the interaction between TRM and TON in potato,the amino acid sequences of TON1A (At3g55000),TON1B(At3g55005),and TON2 (At5g18580) were blasted against potato proteins and the two best hits were identified. Putative StTON1 (PGSC0003DMP400055610)showed 68% homology to TON1A and 67% to TON1B in protein sequence,and putative StTON2(PGSC0003DMP400029382) showed 91% homology to TON2 in protein sequence (Appendix G). The Y2H assay showed that TON2 exhibited interactions with TRM5,TRM19,and TRM20,whereas TON1 did not interact with any of the three TRM proteins (Fig.6).
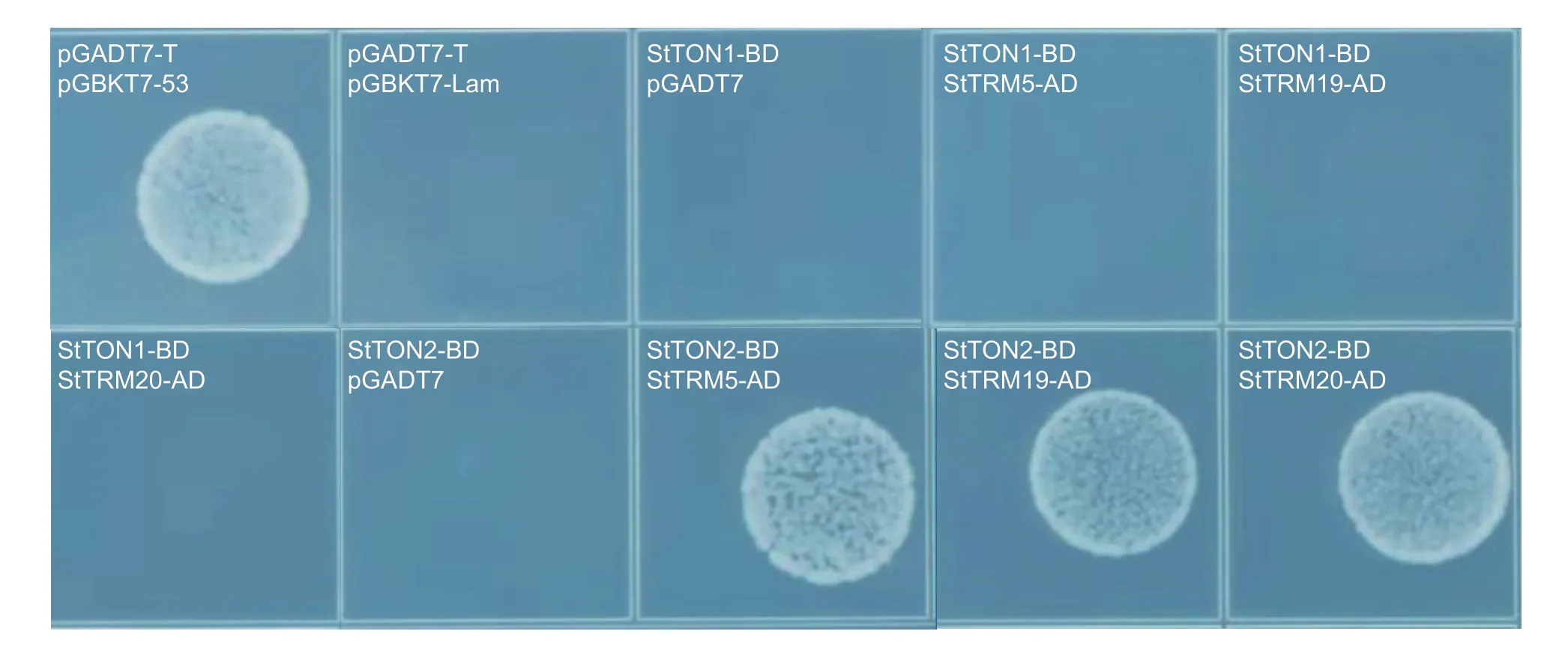
Fig.6 The Y2H assay showing that a putative TON2 interacted with three TRM proteins in potato. pGADT7-T+pGBKT7-53 is the positive control and pGADT7-T+pGBKT7-Lam is the negative control.
4.Discussion
As a plant-specific protein family,OFP members have been shown to control multiple aspects of plant growth and development inArabidopsisand rice (Wanget al.2003,2011;Xiaoet al.2017;Yanget al.2018;Zhouet al.2019;Zhanget al.2020). At the molecular level,they function either as transcriptional repressors or by interacting with various transcription factors. In tomato,SlOFP20 regulates fruit shape as well as floral organ and pollen development by interacting with TRM proteins and mediating hormonal signaling (Wuet al.2018;Zhouet al.2019). This study provided evidence that StOFP20
is expressed in stolons and tubers,and governs potato tuber shape. Compared with its expression in stolons and tubers,StOFP20exhibited substantially stronger expression in flowers suggesting its possible role in flower development. In fact,SlOFP20accumulated high transcript abundance in flowers,and overexpression ofSlOFP20resulted in altered floral architecture and reduced male fertility (Zhouet al.2019).
Both SlOFP20 and StOFP20 showed the maximum protein sequence similarity to AtOFP1 (Appendix H).Functional analysis of AtOFP1 implied that its interaction with TTP components impacted the organization of microtubule arrays which has profound effects on cell division,cell elongation and cell shape (Zhanget al.2020). A Y2H assay identified 11 TRM proteins that interact with SlOFP20 (Wuet al.2018). Because all 11 TRM proteins contain the M10 motif,and TRM proteins without this motif were not recovered,and Wuet al.(2018)speculated that the M10 motif was indispensable for the interactions. This speculation was supported by the fact that substitution of a single amino acid in the M8 motif of three TRMs,as well as in the OVATE domain of SlOFP20,impaired the interaction strength. In our study,among the 23 amplified TRMs,11 proteins harbor the M10 motif and three of them interacted with StOFP20. The three potato TRMs have their corresponding homologs in the 11 tomato TRM proteins,indicative of the overlapping components in their regulatory mechanisms. Mutations of the M8 motif negatively affected the interactions between SlOFP20 and SlTRM5,SlTRM25 and SlTRM17/20a(Wuet al.2018). However,deletion of the M10 motif abolished the interaction between StOFP20 and StTRM5,but it did not exert any effects on the interactions between StOFP20 and StTRM19 and StTRM20. Comparison of the TRM protein sequences revealed that SlTRM5 and StTRM5 share high similarity,and SlTRM17/20a and StTRM20 also possess high similarity,but SlTRM25 and StTRM19 only share quite low similarity (Appendix I).Therefore,sequence dissimilarity may partially explain the differences in the deployment of the M8 or M10 motifs,although other factors may also account for these differences. First,we noticed that even when the same parameter was set for the searches of the M10 or M8 motifs,the interval containing a relatively conserved 10 amino acids is considered as the M8 motif,while the M10 motif is comprised of 14 amino acids. Second,the M8 motif was mutated (Wuet al.2018) and the M10 motif was deleted in this study. Therefore,further investigation is needed to clarify the roles of the M8 or M10 motifs in the interaction. The requirement of the OVATE domain differs among OFPs and its interactants. AtOFP1 regulates the development of multiple organs,and its function requires the involvement of TON2 (Zhanget al.2020). The interaction of AtOFP1 and TON2 was not mediated by the OVATE domain,but in contrast,the OVATE domain was critical for the interactions between OFP20 homologous proteins and TRM proteins (Fig.5-B) (Wuet al.2018).
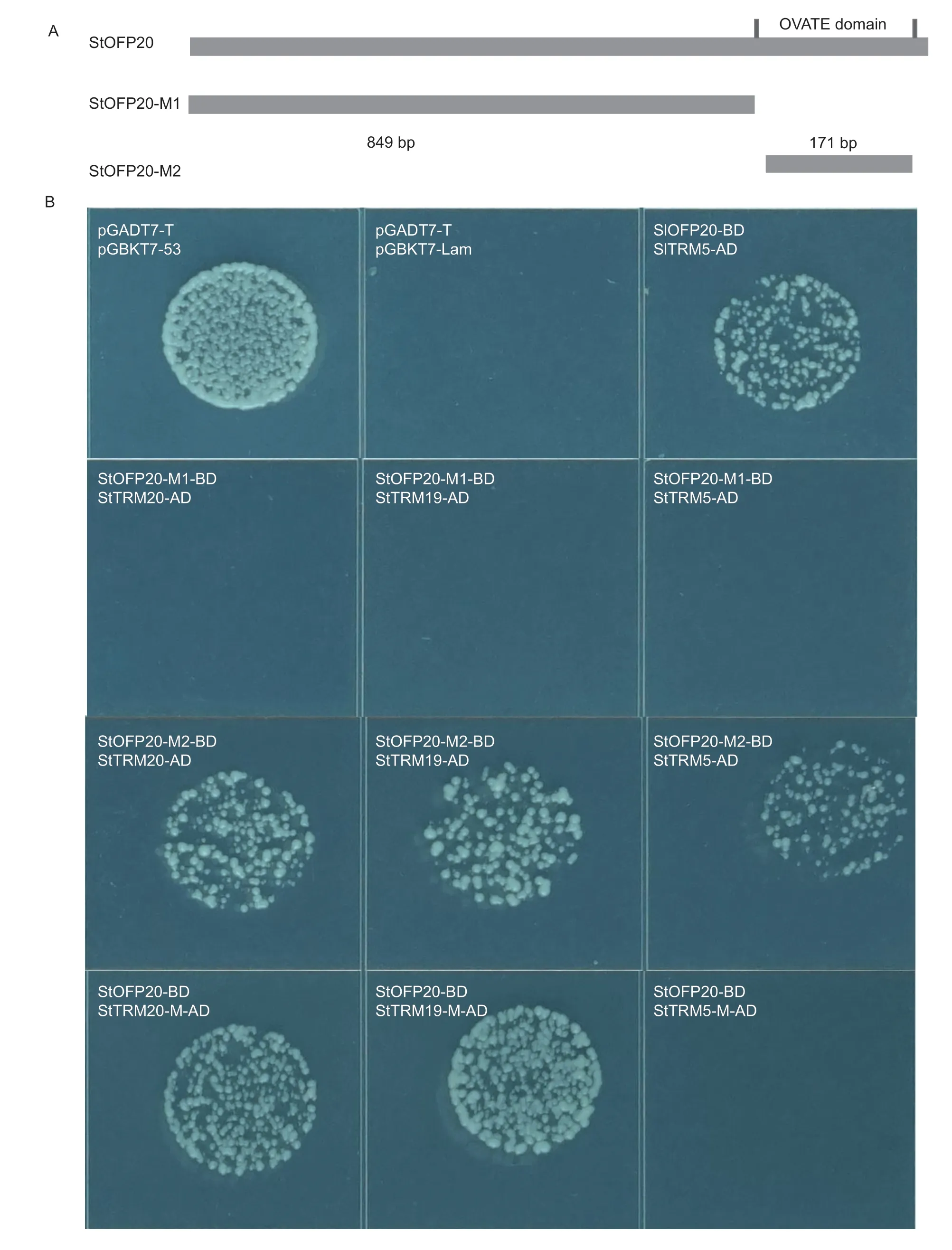
Fig.5 Necessity of the OVATE domain and the M10 motif for the StOFP20 and TRMs interactions. A,schematic of the constructs containing intact OFP20 and fragmented OFP20. pGADT7-T+pGBKT7-53 is the positive control and pGADT7-T+pGBKT7-Lam is the negative control. B,Y2H assay showing that the OVATE domain was necessary for the interactions between OFP20 and TRM proteins,and the interactions responded differently to the deletion of the M10 motif.
Wuet al.(2018) proposed that OFP20-dependent localization of the TRM proteins prevents the availability of TRMs in microtubules,which impairs the recruitment of TTP components to microtubules by TRMs and further compromises the proper function of the TTP complex in PPB formation and cortical array organization(Lazzaroet al.2018;Wuet al.2018). It is noteworthy that the interaction of TRM5 and OVATE only led to the relocalization of TRM5,whereas the interaction of TRM5 and SlOFP20 altered the localization of both proteins (Wuet al.2018). Whether the movement of SlOFP20 to the microtubules has a biological function is an intriguing question. Zhanget al.(2011) showed that AtOFP1 directs the interactions with TON1,TON2 and TRM1. Since the function of TON2 is related to α-tubulin phosphorylation and microtubule nucleation(Kiriket al.2012;Lazzaroet al.2018),one possibility is that the interaction of AtOFP1 with TON2 affects these two processes to modulate microtubule reorientation. In potato,we identified three TRM proteins interacting with StOFP20 and a TON2 protein interacting with the three TRMs. However,the interactions between StOFP20 and TON proteins were not detectable (Appendix J)(Zhanget al.2020). Considering that the interaction of TRM5 and SlOFP20 led to the movement of SlOFP20 to the microtubules,the identification of additional OFP20 interacting proteins,such as proteins involved in microtubule organization processes,will provide further insight into the mechanisms by which OFP20 modulates harvestable organ growth.
5.Conclusion
StOFP20underlies theRolocus and its mutation is responsible for the conversion of potato tubers from round to oval shapes. Y2H screening identified three TRM proteins that interact with StOFP20.The requirement of the conserved OVATE domain was universal,while the absence of the M10 motif had different effects on the interactions. Our findings revealed that OFP20 homologs play a common role in regulating harvestable organ growth,and they may employ partially overlapping mechanisms in the regulatory network.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32060684) and the Academician Workstation of Yunnan,China (202105AF150028).
Declaration of competing interest
The authors declare that they have no conflict of interests.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
 Journal of Integrative Agriculture2023年3期
Journal of Integrative Agriculture2023年3期
- Journal of Integrative Agriculture的其它文章
- Germinated brown rice relieves hyperlipidemia by alleviating gut microbiota dysbiosis
- Melatonin treatment alleviates chilling injury in mango fruit 'Keitt'by modulating proline metabolism under chilling stress
- Changes in the activities of key enzymes and the abundance of functional genes involved in nitrogen transformation in rice rhizosphere soil under different aerated conditions
- Growth and nitrogen productivity of drip-irrigated winter wheat under different nitrogen fertigation strategies in the North China Plain
- Effect of fertigation frequency on soil nitrogen distribution and tomato yield under alternate partial root-zone drip irrigation
- Phylogenetic and epidemiological characteristics of H9N2 avian influenza viruses in Shandong Province,China from 2019 to 2021
