Fluorescent aptasensor for detection of live foodborne pathogens based on multicolor perovskite-quantum-dot-encoded DNA probes and dual-stirring-bar-assisted signal amplification
Liu Liu ,Juncheng Hong ,Wenhi Wng ,Shu Xio ,Hongzhen Xie ,Qiqin Wng ,Ning Gn ,*
a School of Material Science and Chemical Engineering,Ningbo University,Ningbo,Zhejiang,315211,China
b Institute of Pharmaceutical Analysis,College of Pharmacy,Jinan University,Guangzhou,510632,China
Keywords:Aptasensor Live bacteria Multiple detection Multicolor perovskite quantum dots Dual-stirring-bar
ABSTRACT In this study,a fluorescent(FL)aptasensor was developed for on-site detection of live Salmonella typhimurium(S.T.)and Vibrio parahaemolyticus(V.P.).Complementary DNA(cDNA)of aptamer(Apt)-functionalized multicolor polyhedral oligomeric silsesquioxane-perovskite quantum dots(cDNA-POSSPQDs)were used as encoded probes and combined with dual-stirring-bar-assisted signal amplification for pathogen quantification.In this system,bar 1 was labeled with the S.T.and V.P.Apts,and then bar 2 was functionalized with cDNA-POSS-PQDs.When S.T.and V.P.were introduced,pathogen-Apt complexes would form and be released into the supernatant from bar 1.Under agitation,the two complexes reached bar 2 and subsequently reacted with cDNA-POSS-PQDs,which were immobilized on MXene.Then,the encoded probes would be detached from bar 2 to generate FL signals in the supernatant.Notably,the pathogens can resume their free state and initiate next cycle.They swim between the two bars,and the FL signals can be gradually enhanced to maximum after several cycles.The FL signals from released encoded probes can be used to detect the analytes.In particular,live pathogens can be distinguished from dead ones by using an assay.The detection limits and linear range for S.T.and V.P.were 30 and 10 CFU/mL and 102-106 CFU/mL,respectively.Therefore,this assay has broad application potential for simultaneous on-site detection of various live pathogenic bacteria in water.
1.Introduction
A report from the World Health Organization shows that about 600 million people are injured byVibrio-polluted foods every year[1].In low-income and middle-income countries,unsafe food causes US$110 billion in economic losses each year[2].Vibrio parahaemolyticus(V.P.),Staphylococcus aureus(S.T.),or other foodborne pathogens can cause serious illness and harm public health[3].To date,for the detection of foodborne pathogens,a variety of analytical methods have been proposed,including the plate count method[4],genomic polymerase chain reaction technology[5,6],loop-mediated isothermal amplification technology[7,8],and enzyme-linked immunoassay technology[9,10].Although these assays exhibit superior sensitivity and specificity[11-13],some shortcomings cannot be ignored,including complicated pretreatment,time and labor consumption.In addition,such assays only analyze a single foodborne pathogen at a time[14,15].In actual food samples,multiple foodborne pathogens may simultaneously exist in the same sample.Hence,designing an efficient and simultaneous multiple detection platform is necessary for foodborne pathogens.Kim et al.[16]reported microarray analysis of four foodborne pathogens using specific probes prepared by comparative genomics.This method exhibited acceptable sensitivity(100 CFU/mL),but the operation time took 2 h.Moreover,the implementation is complicated,and it requires professional knowledge and technology.Thus,rapid and simultaneous monitoring of multiple foodborne pathogens with simple operation,high sensitivity,and strong specificity remains a challenge.
Recently,a fluorescent(FL)aptasensor has been proposed for multivariate analysis,for example,for pathogens or biomarkers[17],by using multicolor luminescent nanoprobes,including carbon dots[18],rare-earth-based nanomaterials[19],and semiconductor quantum dots[20].To date,nanoprobe-based FL biosensors have become a powerful tool for bioanalysis[21-23].In designing and fabricating multicolor nanoprobes,the particle size of the traditional luminescent material(including II-VI or III-V semiconductor nanocrystals)can affect the emission peak;thus,batch differences in particle size may lead to inconsistent optical properties[24,25].Perovskite quantum dots(PQDs)were normally synthesized by CsPbX3nanocrystals(X=Cl,Br,or I)and regarded as a new type of semiconductor quantum dots.Recently,PQDs have received considerable attention in fabricating biosensor probes because of their distinctive optical properties,such as excellent quantum yield[26],less photobleaching[27],and broad excitation spectrum[28].Compared with traditional quantum dot-based nanoprobes,the emission peak position of CsPbX3nanocrystals has a higher consistency in optical performance because it is primarily determined on the basis of the composition of the halide.In particular,CsPbX3with various colors could be determined by changing the composition of halogen elements in PQDs.Thus,PQDs play a great role in manufacturing multicolor and FL probes for the simultaneous detection of multiple pathogens.
A variety of signal amplification methods assisted by enzymes have been developed to enhance the sensitivity of nanomaterialfunctionalized FL sensors,such as exonuclease amplification[29]and dual-specific endonuclease-mediated sensitization[30].Nevertheless,the enzyme used for amplifying the signal is expensive and vulnerable to environmental factors(temperature,pH,etc.)[31].To overcome these drawbacks,enzyme-free amplified strategies,including catalytic hairpin assembly[32]and hybridization chain reaction[33],have been developed.However,background signals without a target are still found in these enzyme-free amplified systems[13].Recently,our group has developed an enzyme-free amplified strategy,namely,dual-stirring-bar-assisted signal amplification(DSBSA)[13].Among the detection system,capture DNA probes were modified on one stirring bar,and trace DNA probes were modified on the other bar to produce signals[13].The target can move between the two bars and react with the modified DNA probes,and the signal source material will be released into the reaction system continuously,thereby achieving signal amplification and analysis of specific pathogens.Moreover,the target can avoid direct contact between DNA probes and avoid background signals.This previous study only used organic FL dye for signal development,which slightly improved sensitivity.
In this study,S.T.and V.P.were selected as target pathogens.complementary DNA(cDNA)of aptamer(Apt)-functionalized multicolor polyhedral oligomeric silsesquioxane-perovskite quantum dots(cDNA-POSS-PQDs)were designed as encoded probes to achieve simultaneous analysis of multiple foodborne pathogens(Fig.S1).The sensitivity was improved through DSBSA.As shown in Scheme 1,bar 1 was labeled with V.P.and S.T.Apts through DNA hybridization,and then bar 2 was modified with MXenes and cDNA-POSS-PQD-encoded probes.When S.T.and V.P.were added,the corresponding pathogen-Apt complexes would form and then be released into supernatant from bar 1.Under stirring,these complexes reached bar 2,and the Apt of the pathogen could react with cDNA-POSS-PQDs to generate an intensive FL signal in the supernatant.The detached pathogens would resume their free state and initiate the next cycle;therefore,the FL signals could be gradually enhanced to the maximum without adding enzymes after several cycles.The high sensitivity of FL signals from the released encoded probes could be achieved for the detection of targets.Moreover,the anti-interference ability,analytical conditions,and assay specificity were systematically investigated.In particular,the discrimination between live and dead bacteria(V.P.and S.T.)was also explored.Finally,the application potential of the novel method was evaluated by simultaneous detection of several foodborne pathogens in aquatic waters.
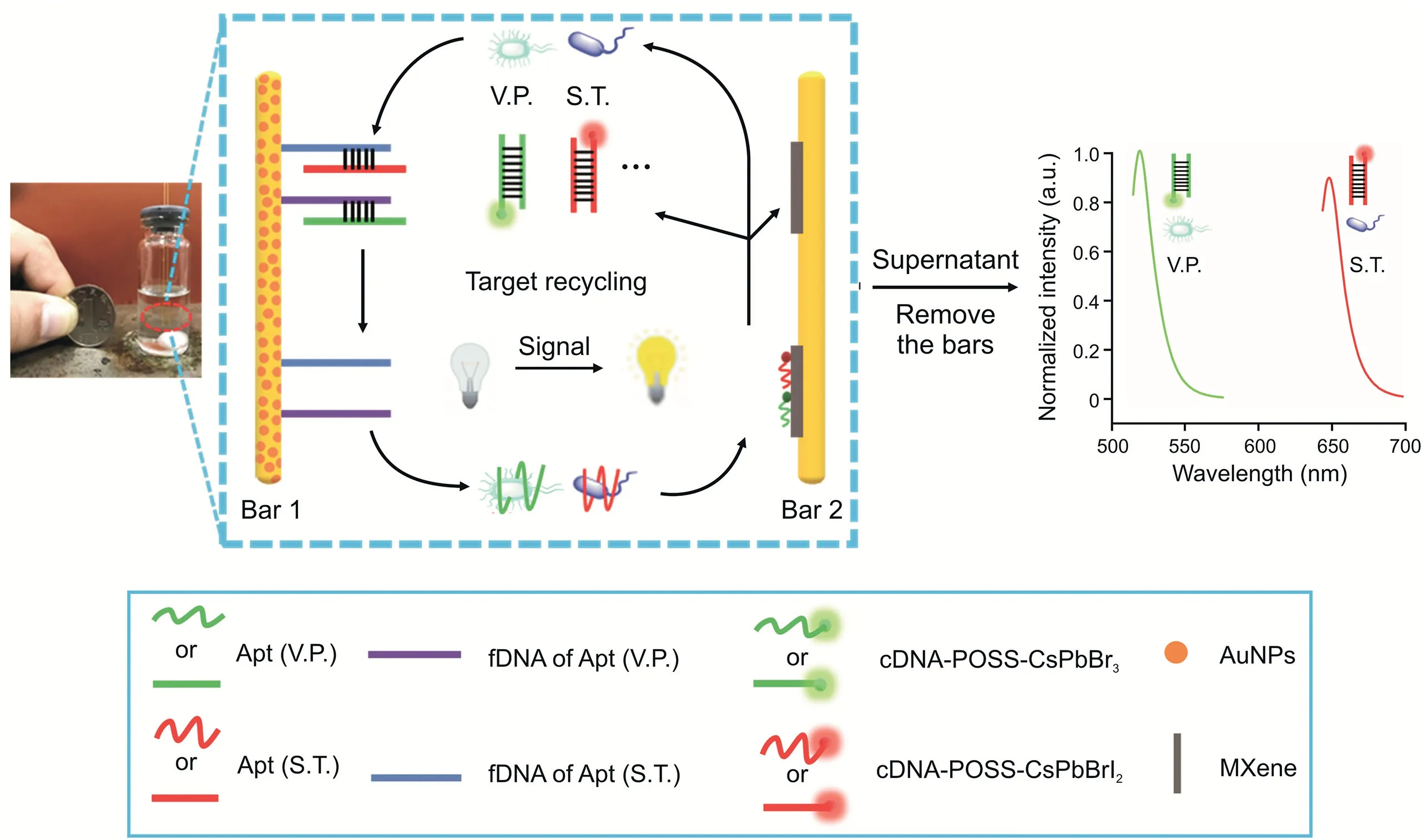
Scheme 1.Schematic illustration of the proposed multicolor fluorescent(FL)aptasensor using a dual-stirring-bar-assisted signal-amplified strategy.V.P.:Vibrio parahaemolyticus;S.T.:Salmonella typhimurium;Apt:aptamer;fDNA:the incomplete complementary strand of Apt;cDNA:complementary DNA of Apt;POSS:polyhedral oligomeric silsesquioxane;AuNPs:Au nanoparticles.
2.Experimental
2.1.Materials and chemicals
PbBr2(99.999%),PbI2(99.999%),Cs2CO3(99.99%),toluene,oleic acid(OA,analytical grade),oleylamine(OAm,80%-90%),octane(99.5%),(3-mercaptopropyl)trimethoxysilane(MPTMS,95%),HAuCl4·3H2O(99.9%,containing 49% Au),tris-ethylenediamine tetraacetic acid buffer solution(TE,pH 8.0),tris(2-carboxyethyl)phosphine hydrochloride(>98%,TCEP),phosphate-buffered saline(PBS,pH 7.4),and sodium citrate(99%)were obtained from Aladdin Reagent Company(Shanghai,China).Gold wire(0.2 mm in diameter)and base sequences of a series of DNA chains(Table S1)were obtained from Sinopharm Chemical Reagent Corporation(Shanghai,China).Ti3AlC2(98%),dimethyl sulfoxide(DMSO),and LiF(99.99%)were purchased from Macklin Co.,Ltd.(Shanghai,China).Octa-aminophenyI-POSS(POSS-NH2,>98%)was obtained from Hybrid Plastics Co.,Ltd.(Hattiesburg,MS,USA).V.P.(ATCC 17802),Staphylococcus aureus(S.aureus,ATCC 43300),Escherichia coli(E.coli,ATCC 25922),and S.T.(ATCC 14028)were provided by Luwei Technology Co.,Ltd.(Shanghai,China).Listeria monocytogenes(CICC 21662)was obtained from the China Center of Industrial Culture Collection(Beijing,China).
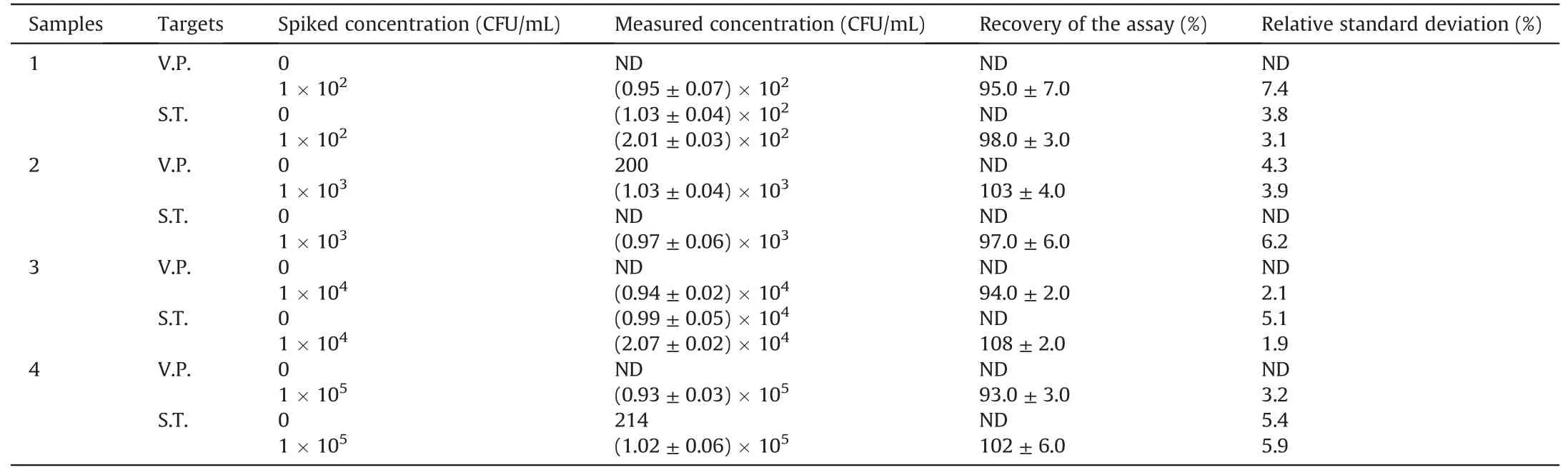
Table 1Real aquatic water samples detected by the proposed method(n=5).
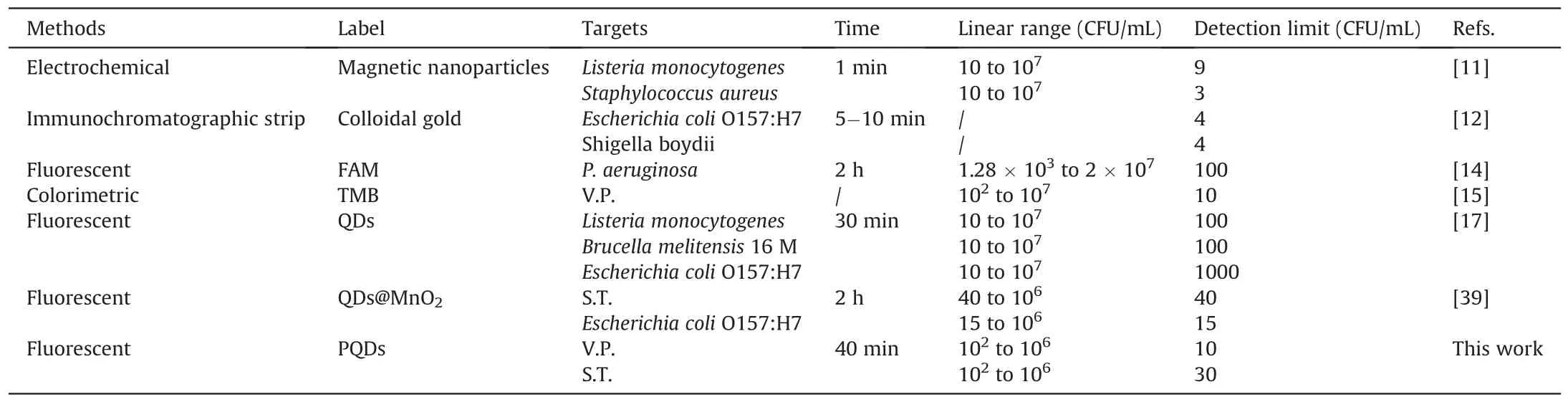
Table 2 Comparison of this novel strategy with other methods for the detection of foodborne pathogens.
2.2.Instruments
An RF-6000 fluorescence spectrometer provided by Shimadzu Co.,Ltd.(Osaka,Japan)was used for fluorescence experiments.A scanning electron microscope with S3400 N mode(Hitachi,Tokyo,Japan)and a transmission electron microscope with H600 mode(Hitachi,Tokyo,Japan)were utilized to explore surface morphology.Surface structure observation was performed on a Bruker's multimode-8 atomic force microscope(Karlsruhe,Germany).A Bruker's D8 Advance X-Ray powder diffractometer(Karlsruhe,Germany)was used to measure X-ray diffraction spectra.Malvern's Zetasizer Nano ZS90(London,UK)was used for dynamic light scattering(DLS)analysis.
2.3.Synthesis of Au nanoparticles(AuNPs)
Based on previous reports,AuNPs were synthesized by citrate reduction[13].Fifty milliliters of 0.1 mg/mL of HAuCl4was initially heated to boiling,and then 0.75 mL of sodium citrate solution(1.0%,m/V)was quickly added to the system.After stirring for 10 min,the mixture was turned from gray to red and finally cooled to room temperature for further applications.
2.4.Synthesis of multicolor CsPbX3 PQDs
Multicolor PQDs were prepared as follows.First,0.6 mmol(195.5 mg)of Cs2CO3powder,1.5 mL of OA,and 30 mL of octane were dropwise mixed in a 100 mL round-bottom flask.Under stirring(500 r/min),the mixture was dissolved and kept at 90°C to obtain Cs precursors.Then,1.8 mmol(0.66 g)of PbBr2,3.5 mL of OAm,and 1.5 mL of OA were added to 30 mL of octane and placed in the abovementioned flask.Under stirring(500 r/min),the mixture was heated at 80°C for 30 min to obtain PbBr2precursors.Afterward,30 mL of PbBr2precursors and 30 mL of Cs precursors were added to a 100 mL round-bottom flask.It was heated at 80°C for 0.25 h.Then,the mixture was placed in an ice-water bath and kept for 10 min.The reactant was centrifuged at 8,000 r/min for 5 min.Next,a rotating speed of 12,000 r/min was set to centrifuge the supernatant for 3 min.Finally,the precipitate was mixed in 2.0 mL ofn-hexane.
A similar process was used for the preparation of CsPbBrI2PQDs.0.6 mmol(0.22 g)of PbBr2,0.4 mL of OA,and 0.8 mL of OAm were dissolved in 10 mL of octane and placed in the 100 mL roundbottom flask.Under stirring(600 r/min),the mixture was heated at 80°C for 5 min to achieve PbBr2precursors.1.8 mmol(0.65 g)of PbI2,2 mL of OAm,and 1.2 mL of OA were dissolved in 25 mL octane,placed in the 100 mL flask,and then kept at 90°C at a rotating speed of 650 r/min for 30 min.Afterward,10 mL of PbBr2precursors,20 mL of PbI2precursors,and 35 mL of Cs2CO3solution were dropwise added to the abovementioned flask and heated at 80°C for 20 min.Then,this mixture was immersed in a container containing 300 mL of ice-water bath for 0.25 h.The resulting mixture was centrifuged at a speed of 8,000 r/min for 5 min.The precipitate was then collected by centrifuging the supernatant at 12,000 r/min and mixed into 5 mL ofn-hexane for the following applications.
2.5.Preparation of cDNA-POSS-PQDs
First,1.2 g of POSS-NH2was added into 10 mL of 45 mg/mL CsPbX3PQDs in toluene and fully dissolved.The abovementioned solution was then heated at 60°C for 0.5 h.The toluene was evaporated in a rotary evaluator to obtain POSS-PQD powders.Next,20 mg of POSS-PQDs was added into 10 mL of 2.5 mM ferrocene solution and stirred for 12 h.Then,the mixture was centrifuged at 12,000 r/min,and the precipitate was washed with 10.0 mL of PBS three times.The precipitate was mixed with 5.0%glutaraldehyde in 5.0 mL of PBS and reacted for 4 h.The products were obtained after being washed with 10.0 mL of PBS three times and then centrifuged at the abovementioned rotating rate.Afterward,20μL of 2.0×10-5M amino-functionalized DNA was reacted with the abovementioned probes and then dissolved in a 5 mL tube for 2.5 h(Fig.S1).Finally,the free DNA was washed away using 5.0 mL of PBS three times.Then,the DNA-labeled probes were obtained after being centrifuged at 10,000 r/min.
2.6.Synthesis of Ti3C2 MXene
The synthesis procedure was based on the literature[34]with some revision.Two gram of LiF was dropwise dissolved into 15 mL of 5.0 M HCl with a stirring rate of 600 r/min.Then,2.0 g of Ti3AlC2was mixed in the abovementioned system and heated at 37°C for 12 h.Afterward,under 5,000 r/min,the precipitation was obtained and then washed with 10.0 mL of H2O for 20 min five times.The pellet was obtained through centrifugation for 8 min at a speed of 4,000 r/min.The product(Ti3C2Tx)was placed under a 60°C vacuum condition and kept for 18 h.Then,Ti3C2Txwas added in 8.0 mL of DMSO and continuously stirred for 12 h.The mixture was centrifuged at 5,000 r/min for 10 min,and the supernatant was discarded.Finally,the as-prepared 2.2 g intercalated Ti3C2Txwas dispersed in 100 mL of deionized water and sonicated for 10 h under N2.The solution was dried at 80°C to obtain the final Ti3C2MXene.
2.7.Preparation of a dual-stirring-bar system
Dual-stirring bars(0.2 mm in diameter and 5 cm in length)were immersed in piranha solution,ultrasonicated for 20 s,rinsed with deionized water,and dried with nitrogen.The abovementioned steps were repeated three to four times to thoroughly clean the stirring surface.The cleaned stirring bar was immersed in MPTMS,kept at room temperature for 4 h,and washed with deionized water five to six times to remove the physically adsorbed MPTMS.
During the preparation of stirring bar 1,the stirring bar obtained in the previous step was placed in AuNP solution and kept at 4°C for 24 h.Both thiol-modified DNA strands(incomplete complementary of the Apt of V.P.and S.T.)were diluted with TE buffer(pH=8.0)to 100μM and stored at 4°C.Then,thiol-modified DNA(200μL,6μM)treated with TCEP(10μM)was added into PBS(200μL).The modified stirring bar was immersed in the abovementioned mixture and then kept at 4°C for 12 h.Afterward,the unbound DNA was washed away by deionized water from the stirring bar.Finally,the stirring bar was immersed in the solution of the two Apt DNA strands of V.P.and S.T.(both are 25 nM)for 2 h.
During the preparation of stirring bar 2,the stirring bar obtained from MPTMS treatment was immersed in 2 mL of 50μg/mL MXene solution,kept at 4°C for 8 h,and then rinsed with deionized water to remove unbound MXene.The MXene-modified stirring bar was inserted into 2 mL of 40μg/mL cDNA-POSS-PQDs and kept at room temperature for 2 h to ensure that cDNA-POSS-PQDs were fully adsorbed on MXene.Finally,the obtained bar 2 was gently rinsed with deionized water to remove unbound cDNA-POSS-PQDs.
2.8.Cultivation of bacteria
Vibriostrains were cultivated in Luria-Bertani at 37°C for 12 h,and the cultivated bacteria were obtained during precipitation by centrifuging at 5,000 r/min for 8 min.The precipitate was then washed three times with 5.0 mL of Tris-HCl(50 mM,pH 7.4),which contained NaCl(120 mM),MgCl2(1.2 mM),and KCl(5.0 mM).Then,the bacteria with different concentrations were stepwise diluted by PBS 10-fold.
2.9.Fluorescence intensity measurement
The solution containing different concentrations of V.P.,S.T.,and 4 mL of PBS(pH 7.4)was added into the designed detection system and reacted at 140 r/min for 40 min.The stirring bars were then removed to measure the fluorescence of the mixed solution.Fluorescence intensity from 519 to 647 nm emission was measured at 365 nm excitation.
2.10.Measurement of foodborne pathogens in real aquatic waters
Based on the GB 4789.7-2013 of the National Health Commission of the People's Republic of China[35],real aquatic samples were initially collected in sterile 250 mL glassy containers(the water sample should not be stored more than 4 h from collection to inspection at 4°C).A sterile pipette was utilized to draw a 10 mL of water sample,which was placed in a conical bottle containing 90 mL of sterile water and some glass beads.The sample was fully shaken to make the mixture uniform and make the bacteria as single as possible.Then,the sample solution was inserted into the solution containing stir bars for testing.The solution contained different concentrations of V.P.,S.T.,and 4 mL of PBS and reacted at 140 r/min for 40 min.The stirring bars were then removed to measure the fluorescence of the mixed solution.The pathogen experiment was reviewed and approved by the Ethics Committee for Animal Experiments of Ningbo University,China(Approved No.:2021-0096).
3.Results and discussion
3.1.Characterization of PQDs,POSS-PQDs,cDNA-POSS-PQDs,AuNP-stirring bar 1,MXene,and MXene-stirring bar 2
A series of measurements of the synthesized PQDs and POSSPQDs was performed and compared to evaluate the influence of POSS coating on PQDs.Scanning electron microscopic(SEM)images show that CsPbBr3and CsPbBrI2nanocrystals on the silicon wafer were in the shape of dense nanoparticles(Fig.S2).Transmission electron microscopic(TEM)images indicate that CsPbBr3PQDs were in cube shape(Fig.1A),and the average side length of the cube was 10±3 nm.The high-resolution TEM image exhibits a lattice gap of 0.58 nm in the darker part,which is consistent with the(110)planes of orthorhombic CsPbBr3(Fig.1B).The powder Xray diffraction(PXRD)pattern of CsPbBr3PQDs is shown in Fig.S3A,and its characteristic peaks at 2θwere 15.1°,21.4°,30.5°,33.9°,37.7°,and 43.5°,corresponding to(100),(110),(200),(210),(211),and(202)reflections of crystal planes.In addition,Fig.S3B shows the PXRD pattern of CsPbBrI2PQDs.These results indicated that two different types of PQDs were synthesized with good crystal forms.Moreover,the SEM image of POSS-CsPbBr3indicated that the size of the obtained material after coating with POSS was approximately 45±5 nm(Fig.1C),which was consistent with the measured value of DLS(Fig.1C inset).Similarly,the particle size of the obtained POSS-CsPbBrI2was approximately 45 nm(Fig.S4).Fig.1D shows the fluorescence spectra of CsPbBr3(red)and CsPbBrI2(black)and ultraviolet(UV)-visible absorption spectrum of MXene.The positions of fluorescence emission peaks of quantum dots were within the ultraviolet absorption range of MXene;thus,the fluorescence quenching phenomenon based on fluorescence resonance energy transfer was well founded.Subsequently,the changes in fluorescence intensity of the resulting POSS-PQDs materials were also monitored before(in black)and after encapsulation(in red).As shown in Fig.S5,the changes in fluorescence intensity were only 10%,which indicated that POSS coating did not affect its optical performance.In particular,the water resistance test showed that no significant change in the fluorescence intensity was observed for POSS-PQDs after immersing in water from 0.1 to 72 h(Fig.S6).Therefore,POSS can not only improve the stability and water resistance of PQDs,but also promote optical transparency to maintain the optical performance of PQDs after packaging.Finally,DNA was successfully modified to POSS-PQDs,and the obtained cDNA-POSS-PQDs were evidenced by the zeta potential change of POSS-PQDs(positively charged)and cDNA-POSSPQDs(negatively charged)(Fig.S7).The SEM image indicated that AuNPs were successfully modified on stirring bar 1(Fig.S8).The TEM image showed that the Ti3AlC2MAX-phase was etched by LiF/concentrated HCl and then intercalated by DMSO.After ultrasonication,the Ti3AlC2MAX-phase was successfully peeled off into a single layer of Ti3C2MXene(Fig.1E).The atomic force microscopic image revealed that the thickness of the monolayer Ti3C2MXene was 1.2-1.6 nm(Fig.1F).In addition,Fig.1G shows the changes in the Fourier transform infrared(FT-IR)spectrum of Ti3AlC2before and after etching.The FT-IR spectrum of MXene showed a strong absorption peak at 3436 cm-1,which might be due to the production of-OH or-F on the surface of the nanosheets after etching.Moreover,the PXRD pattern(Fig.S9)shows that the strongest(104)peak of Ti3AlC2(2θ=39°)was significantly weakened after the Ti3AlC2was etched,and the(002)peak of Ti3AlC2broadened and shifted toward a lower angle.These results indicated the transformation of Ti3AlC2to Ti3C2with the removal of Al layers,which was consistent with the reported literature[36].The SEM image further shows the morphology of MXene on the stirring bar,indicating that MXene was successfully modified on stirring bar 2(Fig.1H).Moreover,a strong fluorescence could still be emitted under a 365 nm UV lamp(Fig.1I).
3.2.Feasibility of the aptasensor
In verifying the feasibility of the designed FL aptasensor for simultaneous analysis of different pathogens and the DSBSA system,the fluorescence spectra of cDNA-POSS-CsPbBr3(red,519 nm)and cDNA-POSS-CsPbBrI2(black,647 nm)are shown in Fig.2.When 103CFU/mL of V.P.and S.T.were mixed in the system,strong FL signals could be detected at 519 and 647 nm,respectively(Fig.2A).In particular,when only 103CFU/mL V.P.was mixed in the system,a strong FL signal was detected at 519 nm,and the FL signal at 647 nm can be neglected(Fig.2B).The reverse phenomenon was observed for 103CFU/mL S.T.(Fig.2C).Therefore,these results indicated that the cross-reaction was negligible among different targets,and the sensor can be applied as a new potential platform for simultaneous analysis of various pathogens in aquatic samples.
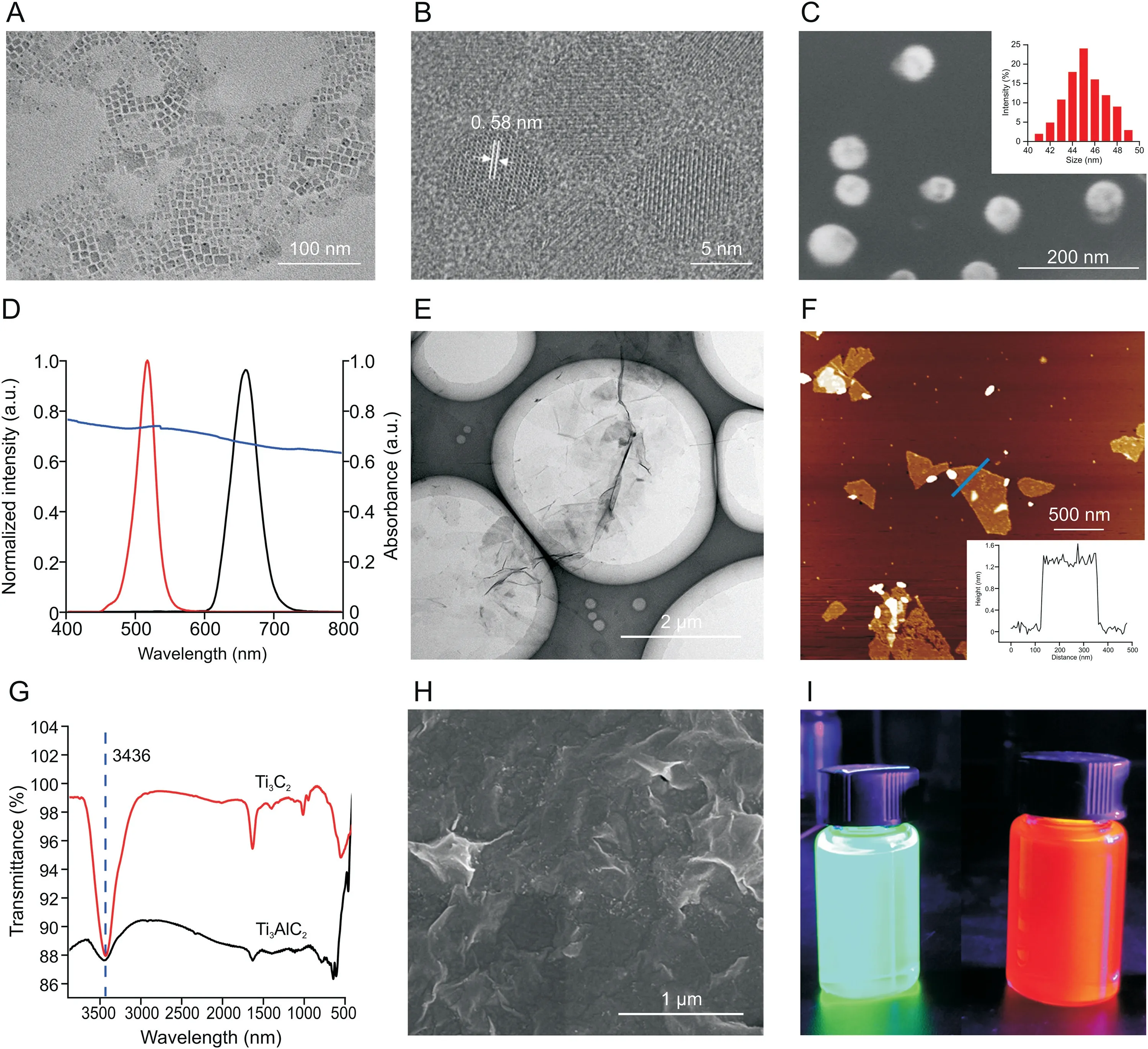
Fig.1.Characterization of perovskite quantum dots(PQDs),polyhedral oligomeric silsesquioxane(POSS)-PQDs,MXene,Ti3AlC2,and MXene on stirring bar 2.(A)Transmission electron microscopy(TEM)image and(B)high-resolution TEM image of CsPbBr3 PQDs.(C)Scanning electron microscopy(SEM)image of POSS-CsPbBr3.Inset:measured dynamic light scattering(DLS)value of POSS-CsPbBr3.(D)Fluorescence spectra of CsPbBr3(red)and CsPbBrI2(black)and ultraviolet(UV)absorption spectra of MXene(purple).(E)TEM image of Ti3C2 nanosheets.(F)Atomic force microscopy image of monolayer Ti3C2 nanosheets.(G)Changes in the Fourier transform infrared(FT-IR)spectrum of Ti3AlC2 before(black)and after(red)etching.(H)SEM image of MXene on stirring bar 2.(I)Real picture of POSS-CsPbBr3(left)and POSS-CsPbBrI2(right)dispersed in water for 72 h under 365 nm UV light.
In addition,the importance of the DSBSA system was investigated.103CFU/mL of S.T.and V.P.was added into the detection aptasensor with and without dual-stirring-bar-assisted target cycling.As shown in Figs.2D,10-fold stronger signals were observed for the two target bacteria in the novel system.This result may be attributed to the fact that the target bacteria-Apt complex formed from bar 1 could react with the cDNA on bar 2,and the target was released into the detection system again for next cycle.These results demonstrated that signal amplification had taken effects.
3.3.Detection mechanism
Microchip electrophoresis was used to investigate the detection mechanism.As shown in Fig.3A,lane 1 and lane 2 represent V.P.’s Apt(Apt[V.P.])and the incomplete complementary strand(fixed DNA(fDNA))of the Apt immobilized on bar 1,respectively.In lane 3,the two strands in lane 1 and lane 2 were mixed.A new band appeared in lane 3,and the original two bands became shallower,which indicated that the Apt combined with fDNA on bar 1 to form double strands.Then,V.P.was added into the system of lane 3,and the double-stranded band became significantly shallower.In addition,the color of the Apt and fDNA bands became darker(lane 4),which indicated that the Apt was pulled from the double strands by V.P.,and the bacteria and Apt were combined and stirred in liquid.Afterward,lanes 5,6,and 7 indicated the mechanism on bar 2.Lane 5 shows the band of the V.P.-Apt complex.Lane 6 shows the cDNA-POSS-CsPbBr3band.Lane 7 is a mixture of two bands.Moreover,a new band was formed,and the bands of the V.P.-Apt complex and individual cDNA-POSS-CsPbBr3were lighter,which indicated that the Apts and corresponding cDNA-POSS-CsPbBr3were hybrid,and double-stranded Apt-cDNA-POSS-CsPbBr3was released into the solution.This finding indicated that dual-stirringbar-assisted target recycling could be achieved for V.P.The verification of the detection mechanism of S.T.by the amplified system is shown in Fig.3B.It had the same effects,demonstrating that S.T.-based target recycling could occur between stirring bar 1 and bar 2.
The reasons why the bacteria-Apt complex could be destructed and form Apt-cDNA-POSS-CsPbBr3on bar 2 were explained by using the following theory calculation.Assuming that one Apt can combine with one V.P.,the dissociation of the V.P.-Apt complex to form V.P.and Apt is demonstrated using Eq.(1).
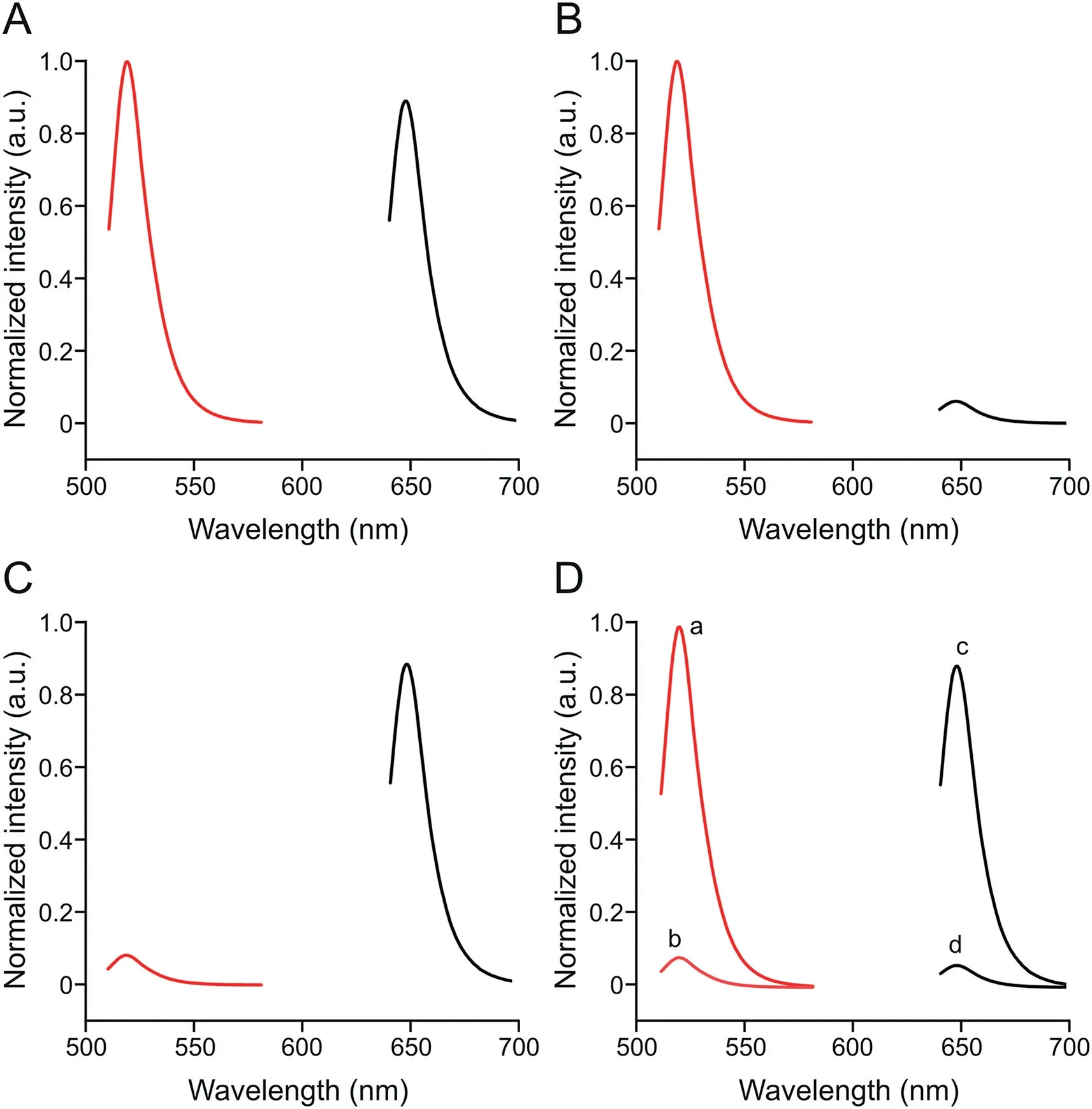
Fig.2.Fluorescence spectra of complementary DNA(cDNA)-polyhedral oligomeric silsesquioxane(POSS)-CsPbBr3(red)and cDNA-POSS-CsPbBrI2(black).(A)103 CFU/mL Vibrio parahaemolyticus(V.P.)and 103 CFU/mL Salmonella typhimurium(S.T.)were added to the reaction system.(B)103 CFU/mL V.P.was added to the reaction system.(C)103 CFU/mL S.T.was added to the reaction system.(D)103 CFU/mL V.P.was added to the reaction system(a);103 CFU/mL V.P.was added to the system without dualstirring-bar-assisted target recycling(b);103 CFU/mL S.T.was added to the reaction system(c);and 103 CFU/mL S.T.was added to the system without dual-stirring-barassisted target recycling(d).

Apt can hybrid with cDNA-POSS-CsPbBr3,and the reaction is shown in Eq.(2).
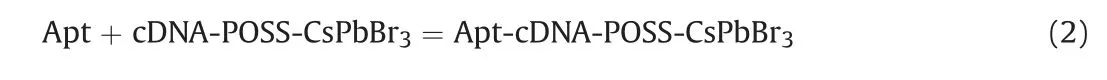
Moreover,the reaction that occurred between V.P.-Apt and cDNA-POSS-CsPbBr3is depicted in Eq.(3).
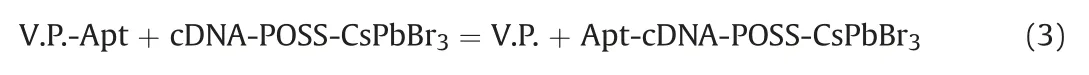
The reaction that occurred between S.T.-Apt and cDNA-POSSCsPbBrI2is depicted in Eq.(4).
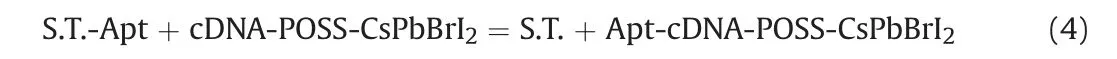
The equilibrium constants of reactions 1 to 3 areK1,K2,andK3,respectively.In addition,the dissociation constant(KD)was equal toK1;therefore,K3=K1×K2=KD×K2.Forty-five hybrid bases were identified between Apt and cDNA-POSS-CsPbBr3.Using the integrated DNA Technologies(IDT)'s calculation software(http://www.idtdna.com/analyzer/applications/oligoanalyz er/),ΔG2was-80.73 kcal/mol.In addition,ΔG=-2.303RTlgK(T=298 K,R=8.315).Therefore,K2was approximately 2.51×1059M-1.Duan et al.[37]reported thatKDof Apt toward V.P.was 16.88 nM.Therefore,K3was approximately 4.24×1051M-1,andΔG3was-295 kJ/mol,which was less than 0.This result indicated that cDNA-POSS-CsPbBr3could react with V.P.-Apt to form Apt-cDNA-POSS-CsPbBr3.ΔG4was also calculated using Eq.(4).Moreover,KDof Apt toward S.T.was 10 nM[38].Another 45 hybrid bases were found between Apt of S.T.and its cDNA-POSS-CsPbBrI2.Therefore,K4was approximately 2.51×1060M-1,andΔG4was-345 kJ/mol,which was less than 0.Therefore,S.T.could also be released from S.T.-Apt by the reaction between cDNA-POSS-CsPbBrI2and Apt.
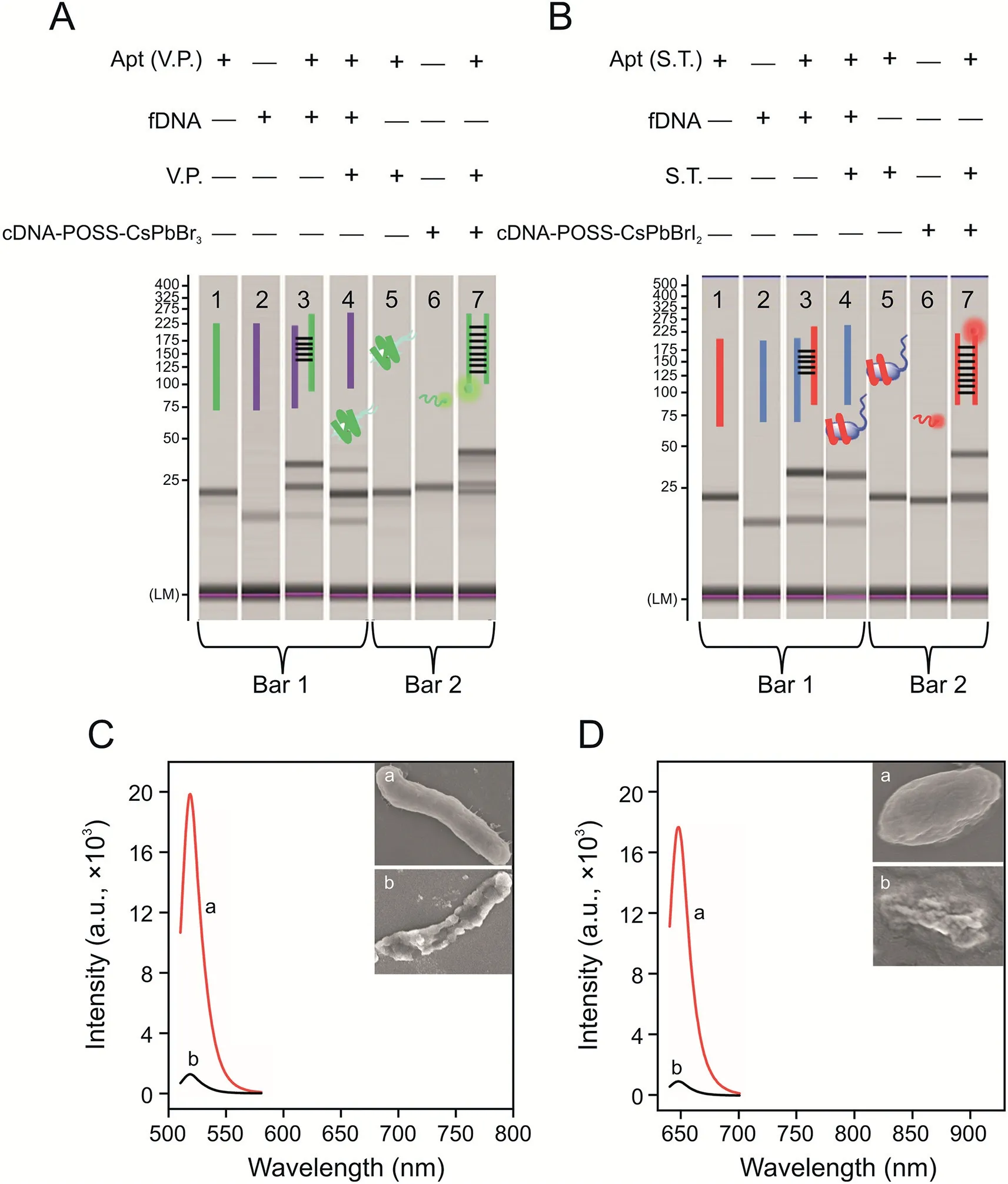
Fig.3.Microchip electrophoresis verification of the dual-stirring-bar-assisted target recycling system of(A)Vibrio parahaemolyticus(V.P.)and(B)Salmonella typhimurium(S.T.)(lane 1:aptamer(Apt)(V.P.);lane 2:the incomplete complementary strand(fixed DNA(fDNA))of the Apt;lane 3:the two strands in lane 1 and lane 2 were mixed;lane 4:the bacteria were added into the system of lane 3;lane 5:bacteria-Apt complex;lane 6:cDNA-POSS-CsPbX3;lane 7:a mixture of bacteria-Apt complex and cDNA-POSSCsPbX3).Comparison of fluorescence spectra of 103 CFU/mL of live and dead bacteria of(C)V.P.(inset:scanning electron microscopy(SEM)images of(a)live and(b)dead V.P.)and(D)S.T.(inset:SEM images of(a)live and(b)dead S.T.).cDNA:complementary DNA of Apt;POSS:polyhedral oligomeric silsesquioxane.
As shown in Figs.3C and D,103CFU/mL of live or dead bacteria(V.P.and S.T.)was added into the system,and the signal intensity of live bacteria was significantly higher(40-fold)than that of bacteria that had been dead for 24 h.Based on inserted SEM images,the live bacteria were intact,whereas the dead bacteria were incompletely broken,which would inevitably lead to a lack of binding sites on the surface of the Apt.Therefore,this system can distinguish and primarily detect live bacteria.
3.4.Optimization of detection conditions
Some key factors,such as the reaction temperature,pH,stirring speed,incubation time,amount of modified MXene,and amount of adsorbed cDNA-POSS-PQDs,were systemically optimized to improve the performance of the proposed sensor.As shown in Fig.4A,when 103CFU/mL of V.P.and 103CFU/mL of S.T.were added into the reaction system,the fluorescence intensity increased with the increase of temperature,and the cDNA-POSSCsPbBr3signal reached the maximum fluorescence intensity at 30°C(the black bar in Fig.4A).However,the signal of cDNA-POSSCsPbBrI2reached the maximum fluorescence intensity at 37°C(the red bar in Fig.4A).This result indicated that the best detection temperature for V.P.was 30°C,whereas the best detection temperature for S.T.was 37°C.The best detection temperature was finally confirmed as 37°C to maximize the fluorescence intensity of the system.Fig.4B shows that the best detection pH for target analytes is 7.0.The speed of stirring bars was optimized,and the highest signal was obtained at 140 r/min(Fig.4C).At 37°C and pH 7.0,103CFU/mL of V.P.was mixed into the solution,and the FL signal of the system reached the maximum in only 40 min(Fig.4D),which was faster than that of the standard plate counting method(about 48 h)for foodborne pathogens.Considering that the insufficient modification amount of MXene on stirring bar 2 could affect the related adsorbed amount of cDNAPOSS-PQDs and the sensitivity of the sensor,the modification amount of MXene was further optimized.The stirring bar after MPTMS treatment was immersed in 2 mL of 20,30,40,50,60,or 70μg/mL MXene solution and then in excess cDNA-POSS-PQD(2 mL,80μg/mL)solution to prepare stirring bar 2 for subsequent reactions.The system signal reached the maximum when the concentration of MXene was 50μg/mL(Fig.4E),which indicated that the optimal concentration of MXene was 50μg/mL.Under the optimal concentration of MXene,the MXene-modified stirring bar was immersed into a 2 mL cDNA-POSS-PQDs solution with different concentrations.Fig.4F shows that when cDNAPOSS-PQDs was 40μg/mL,the FL signal reached its maximum value.Therefore,the optimal reaction temperature,pH,stirring speed,reaction time,MXene concentration,and cDNA-POSS-PQDs concentration were 37°C,7.0,140 r/min,40 min,50μg/mL,and 40μg/mL,respectively.

Fig.4.Optimization of detection conditions:(A)reaction temperature,(B)detection pH,(C)stirring speed,(D)incubation time,(E)amount of modified MXene,and(F)amount of adsorbed complementary DNA(cDNA)-polyhedral oligomeric silsesquioxane(POSS)-perovskite quantum dots(PQDs).
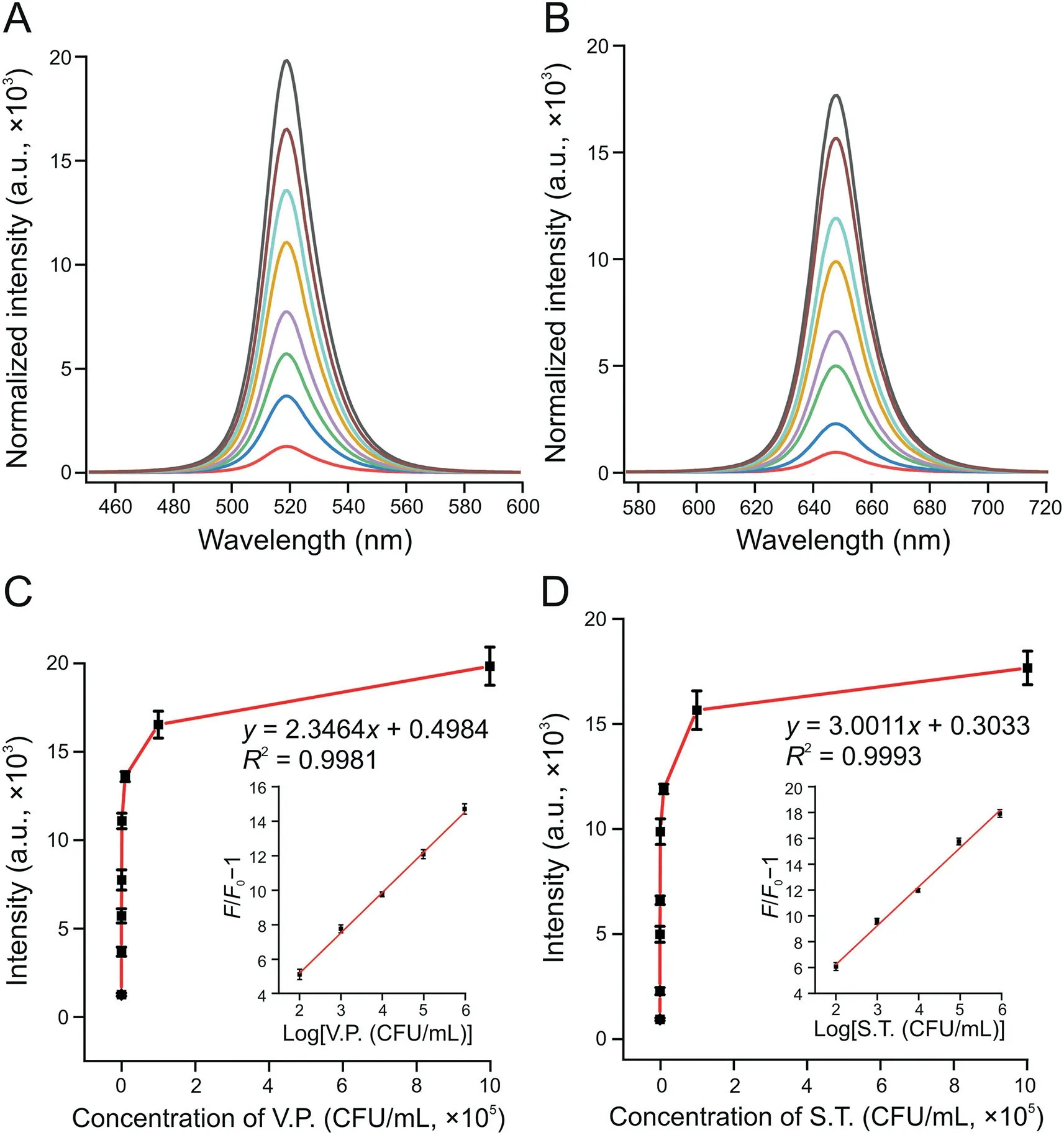
Fig.5.Fluorescence intensity changes of the system at(A)519 nm with Vibrio parahaemolyticus(V.P.)and(B)647 nm in Salmonella typhimurium(S.T.)samples.The curve of the fluorescent(FL)signal changed with the concentrations of(C)V.P.(inset:the relationship between the relative increase of fluorescence and the logarithm of the V.P.concentration)and(D)S.T.(inset:the relationship between the relative increase of fluorescence and the logarithm of the S.T.concentration).The concentrations of V.P.and S.T.from bottom to top in Figs.5A and B are 0,10,50,102,103,104,105,and 106 CFU/mL.The concentrations of V.P.and S.T.from left to right in Figs.5C and D are 0,10,50,102,103,104,105,and 106 CFU/mL.
3.5.Detection performance of the novel sensor
In quantitatively analyzing the analytes,V.P.-and S.T.-spiked water samples were added into the reaction system under optimal working conditions.Figs.5A and B show the changes in fluorescence intensity of the system at 519 and 647 nm in the presence of V.P.and S.T.In addition,as the concentration of targets increased,the FL signal continued to increase(Figs.5A and B).When the V.P.concentration range was between 102and 106CFU/mL,the relationship between the FL signal and logarithm of the V.P.concentration could be described asF/F0-1=2.3464 logc+0.4984,with the coefficient ofR2=0.9981(Fig.5C inset).In the function,F0andFrepresented the FL intensity at 519 nm without and with V.P,respectively.Similarly,when the concentration of S.T.ranged from 102to 106CFU/mL,the relationship between the FL signal and the logarithm of the S.T.concentration could be described asF/F0-1=3.0011 logc+0.3033,with the coefficient ofR2=0.9993(Fig.5D inset).Figs.5C and D show that the limits of detection(LODs)of V.P.and S.T.were 10 CFU/mL(S/N=3)and 30 CFU/mL(S/N=3),respectively.
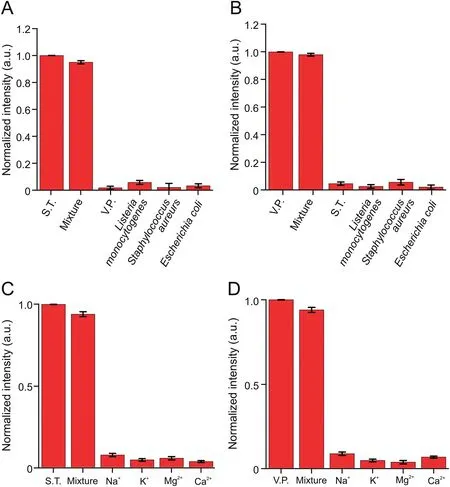
Fig.6.Several common bacteria were added to the reaction system,including(A)Vibrio parahaemolyticus(V.P.)and(B)Salmonella typhimurium(S.T.).The influence of common ions on the test results,(C)S.T.and(D)V.P.Mixture:a mixed sample of test substances shown in a single sheet.
The food sample matrix is complex;thus,other foodborne pathogens and various interferences could cause false-positive signals in the practical application of the sensor.Several common bacteria were added into the reaction system to evaluate the specificity and anti-interference ability of the novel sensor,includingListeria monocytogenes,E.coli,andS.aureus(105CFU/mL)compared with V.P.and S.T.(at a concentration of 103CFU/mL).As shown in Figs.6A and B,when V.P.or S.T.was present,a strong FL signal could be obtained.Bacteria other than V.P.or S.T.did not emit evident FL signals.Moreover,the effect of common ions was studied,including Na+,K+,Mg2+,and Ca2+.The experimental results showed that the influence of these common ions on the test results could be ignored(Figs.6C and D).Furthermore,the strong binding capacity between the target and the Apt ensured that the system exhibited good selectivity and anti-interference ability.
A certain number of S.T.or V.P.were dissolved in the pre-treated aquatic water for detection and calculation of recovery to verify the applicability of this assay to detect real samples.As shown in Table 1,the recovery of V.P.and S.T.in waters was 93%-103% and 97%-108%,respectively.In addition,the relative standard deviation values were 2.1%-7.4% and 1.9%-6.2%,respectively.The excellent performance indicated that this novel platform could simultaneously detect multiple foodborne pathogens in a practical sample.Finally,the novel method was also compared with previously reported methods,such as FL,colorimetric,and immunochromatographic strips(Table 2)[11,12,14,15,17,39].For example,Song et al.[12]proposed an immunochromatographic strip with two kinds of antibodies for screening two foodborne pathogens in cumbersome steps.Eissa et al.[11]reported an electrochemical method based on magnetic nanoparticles,which required the introduction of enzymes,and the process was complicated and environmentally sensitive.Although good detection results can be achieved,these two methods are expensive,and the antibody preparation process is complicated,time consuming,and irregular in quality.Moreover,the FL detection method developed by Gao et al.[14]with carboxyfluorescein as the label and the colorimetric method developed by Sun et al.[15]can detect only one pathogen at a time.Compared with antibodies,quantum dots are cheap and easier to prepare.Moreover,PQDs exhibited a long fluorescence lifetime and good stability,with precise and adjustable emission wavelength compared with traditional organic dye FL probes.Recently,Li et al.[17]developed an FL method with quantum dots to detect pathogenic bacteria with low sensitivity(LODs:102-103CFU/mL).In our study,the Apt-functionalized FL sensor exhibited comparable or better detection performances(easy fabrication,low cost,and good sensitivity)than the above-mentioned methods.This superiority could be attributed to the high stability and selectivity of cDNAPOSS-PQDs as encoded probes and the high efficiency of enzymefree target cycle amplification based on dual-stirring bars.
4.Conclusions
In this study,a multicolor fluorescence assay with high sensitivity and selectivity was fabricated for simultaneous measurement of different live foodborne pathogens.Multicolor quantum dots could simultaneously detect multiple foodborne pathogens in the same system.The dual-stirring-bar system enriched the target to minimize the interference of the matrix and enzyme-free signal amplification.Moreover,the assay could distinguish live bacteria from dead ones.Compared with previously reported methods,the novel method exhibited comparable or better detection performances,including easy fabrication,low cost,and good sensitivity(10 CFU/mL of V.P.and 30 CFU/mL of S.T.).Therefore,this novel platform shows great application potential as a good alternative for the detection of multiple foodborne pathogens in complex matrixes.
CRediT author statement
Liu Liu:Formal analysis,Validation,Investigation,Data curation,Visualization,Writing-Original draft preparation;Juncheng Hong:Methodology,Software,Conceptualization;Wenhai Wang:Data curation,Visualization,Validation;Shu Xiao:Data curation,Validation;Hongzhen Xie:Data curation,Investigation;Qiqin Wang:Writing-Reviewing and Editing,Validation,Investigation;Ning Gan:Conceptualization,Methodology,Resources,Supervision,Writing-Reviewing and Editing,Project administration,Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China(Grant No.:21974074),Ningbo Public Welfare Technology Plan Project of China(Grant Nos.:2021Z056,2022Z170,2022S011,and 202002N3112),Zhejiang Provincial Top Discipline of Biological Engineering(Level A)(Grant Nos.:CX2021051 and KF2021004),Zhejiang Province Public Welfare Technology Application Research Analysis Test Plan(Grant No.:LGC20B 050006),and K.C.Wong Magna Fund in Ningbo University.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2022.07.001.
 Journal of Pharmaceutical Analysis2022年6期
Journal of Pharmaceutical Analysis2022年6期
- Journal of Pharmaceutical Analysis的其它文章
- Corrigendum to“The potential of miRNA-based therapeutics in severe acute respiratory syndrome coronavirus 2(SARS-CoV-2)infection:A review”[J.Pharma.Anal.11(2021)265-271]
- Development of a surface plasmon resonance biosensor for accurate and sensitive quantitation of small molecules in blood samples
- Peptide-RNA complexation-induced fluorescence“turn on”displacement assay for the recognition of small ligands targeting HIV-1 RNA
- Tumor-targeting intravenous lipid emulsion of paclitaxel:Characteristics,stability,toxicity,and toxicokinetics
- Metabolomic and elemental profiling of blood serum in bladder cancer
- A highly efficient protein corona-based proteomic analysis strategy for the discovery of pharmacodynamic biomarkers
