A strategy of screening and binding analysis of bioactive components from traditional Chinese medicine based on surface plasmon resonance biosensor
Diya Lv,Jin Xu,Minyu Qi,Dongyao Wang,Weiheng Xu,Lei Qiu,Yinghua Li,Yan Cao,*
aCenter for Instrumental Analysis,School of Pharmacy,Second Military Medical University,Shanghai,200433,China
bDepartment of Neurology,Changzheng Hospital,Second Military Medical University,Shanghai,200003,China
cDepartment of Biochemical Pharmacy,School of Pharmacy,Second Military Medical University,Shanghai,200433,China
dDepartment of Pharmaceutical Analysis,School of Pharmacy,Second Military Medical University,Shanghai,200433,China
eInstitute of Translational Medicine,Shanghai University,Shanghai,200444,China
ABSTRACT
Elucidating the active components of traditional Chinese medicine(TCM)is essential for understanding the mechanisms of TCM and promote its rational use as well as TCM-derived drug development.Recent studies have shown that surface plasmon resonance(SPR)technology is promising in this field.In the present study,we propose an SPR-based integrated strategy to screen and analyze the major active components of TCM.We used Radix Paeoniae Alba(RPA)as an example to identify the compounds that can account for its anti-inflammatory mechanism via tumor necrosis factor receptor type 1(TNF-R1).First,RPA extraction was analyzed using an SPR-based screening system,and the potential active ingredients were collected,enriched,and identified as paeoniflorin and paeonol.Next,the affinity constants of paeoniflorin and paeonol were determined as 4.9 and 11.8μM,respectively.Then,SPR-based competition assays and molecular docking were performed to show that the two compounds could compete with tumor necrosis factor-α (TNF-α)while binding to the subdomain 1 site of TNF-R1.Finally,in biological assays,the two compounds suppressed cytotoxicity and apoptosis induced by TNF-α in the L929 cell line.These findings prove that SPR technology is a useful tool for determining the active ingredients of TCM at the molecular level and can be used in various aspects of drug development.The SPR-based integrated strategy is reliable and feasible in TCM studies and will shed light on the elucidation of the pharmacological mechanism of TCM and facilitate its modernization.
Keywords:
Surface plasmon resonance
Bioactive components
Screening
Binding analysis
Radix Paeoniae Alba
1.Introduction
Traditional Chinese medicine(TCM)is highly valued in China and has been used widely in Asian and European countries to prevent and cure diseases[1-4].However,there are still some concerns about the efficacy,side effects,and mechanisms of action of TCM due to its complexity,which has hindered its development.In order to gain a better understanding of TCM,it is necessary to elucidate the active components of TCM,their protein targets,and the relationship between compounds and targets at the molecular level.Therefore,investigation of active components of TCM should be the focus of further TCM studies,which will bring TCM in line with modern medicine and pharmacology standards.
Among the methods of investigating into the active components of TCM,surface plasmon resonance(SPR)technology has gained attention in recent years.SPR is a classical tool used in drug discovery and can provide simultaneous kinetic and equilibrium characterizations of molecular interactions[5-8].We and other research groups have established SPR methods to screen active components from TCM and have demonstrated the advantages of SPR in this field[9-12].For example,Zhang et al.[13]described a combination system of SPR and high-performance liquid chromatography-mass spectrometry(HPLC-MS)to identify compounds from Radix Astragali that can bind to human serum albumin.They discovered 20 bioactive components and proved that this method is rapid and possesses a high-throughput.Cao et al.[14]constructed an SPR-based and lentivirus-based membrane protein ligand screening system to identify P-glycoprotein(P-gp)inhibitors from 40 compounds.They discovered three potential P-gp ligands and provided solid evidence that SPR-based screening technology is suitable for complicated protein targets.In the SPR-based active ingredient screening system,ultra-high performance liquid chromatography quadrupole time-offlight mass spectrometry(UPLC-QTOF/MS)was used to identify the active compounds collected from SPR recovery.UPLC-QTOF/MS is a powerful tool for the qualitative and quantitative analyses of TCM components.A variety of UPLC-QTOF/MS applications show that it can achieve ideal separation of multiple components and generate accurate mass information that can be used for identification[15,16].Therefore,the combination of SPR and UPLC-QTOF/MS systems could be used to monitor biomolecular interactions in real time and screen specific components targeted to proteins without destroying their molecular activity,making it a sensitive,efficient,and convenient screening tool for illuminating complex TCM drug systems.
Radix Paeoniae Alba(RPA)is commonly used for its antiinflammatory and immune-regulatory effects[17].From the in vivo aspect,RPA has historically been used as a valuable herb in the treatment of rheumatoid arthritis[18-20]and hepatitis[21,22].From the in vitro aspect,RPA has been reported to inhibit cell proliferation and the production of pro-inflammatory cytokines,such as tumor necrosis factor-α (TNF-α),prostaglandin E2,cyclic adenosine monophosphate,and interleukins-1[23-25].In addition,the anti-inflammatory activities of some ingredients in RPA extracts have been investigated.For example,Bi et al.[26]found that monoterpenoids from RPA have anti-inflammatory effects on lipopolysaccharides-stimulated RAW 264.7 cells.Xie et al.[27]reported that paeoniflorin attenuated hepatic inflammation responses by inhibiting the HMGB1-TLR4 signaling pathway.However,there is little knowledge about the anti-inflammatory mechanism of RPA as well as the potential bioactive components and direct binding target proteins responsible for its pharmacological activities.Because RPA inhibits the production of proinflammatory cytokines,such as TNF-α,which play a key role in the inflammatory response through tumor necrosis factor receptor type 1(TNF-R1)signaling,characterization of active ingredients from RPA that bind to TNF-R1 is needed to understand the potential bioactive components that can account for the anti-inflammatory effects of RPA and how these bioactive candidates initiate signal transduction pathways.
Herein,we propose an integrated strategy based on SPR to screen and analyze the bioactive components of TCM.We used RPA as an example and revealed its components that could act on TNFR1 and account for its anti-inflammatory mechanism.First,an SPR screening system was used to identify the TNF-R1-bound components from RPA;paeoniflorin and paeonol were recovered and identified.The affinity constants of paeoniflorin and paeonol were determined by SPR affinity analysis as 4.9 and 11.8μM,respectively.Then,SPR-based competition assays and molecular docking were performed to show that the two compounds could compete with TNF-α while binding to the subdomain 1 site of TNF-R1.Finally,in cell viability assays,TNF-α and actinomycin D(Act-D)caused nearly 70% cytotoxicity in L929 cells,which was inhibited in a dosedependent manner by paeoniflorin and paeonol.In cell apoptosis analyses,the levels of apoptosis increased markedly to 60.02% upon TNF-α and Act-D treatment,and this induction was suppressed by the two compounds.These results prove that the strategy is reliable and feasible and can provide information on the pharmacological mechanism of TCM and facilitate its modernization development.
2.Materials and methods
2.1.Chemicals and reagents
RPA was obtained from Leiyunshang Pharmacy(Shanghai,China).Paeoniflorin and paeonol were purchased from the National Institute for Pharmaceutical and Biological Products of China(Beijing,China).Act-D was purchased from Sigma-Aldrich(St.Louis,MO,USA).TNF-α and TNF-R1 were purchased from PeproTech(Rocky Hill,NJ,USA).HBS-EP+buffer was purchased from GE Healthcare(Uppsala,Sweden).
2.2.Extraction of RPA
The dry roots of RPA were powdered and weighed.For ultrasound-assisted extraction,powdered herbs(1 g)were suspended in 10 mL of 80% methanol and extracted in an ultrasonic processor for 30 min.The extraction was centrifuged at 12,000 r/min for 10 min,and the supernatant was collected and stored at 4°C.
2.3.Characterization of the sensor chip
A Biacore T200 system(GE Healthcare,Uppsala,Sweden)was used to conduct SPR assays.TNF-R1 and TNF-α proteins were immobilized on CM5 sensor chips using an amine coupling procedure.The TNF-R1 chip was used for screening,validation,and competition assays,whereas the TNF-α chip was only used for competition analysis.Binding specificity was assessed in HBSEP+at a constant flow rate of 30 μL/min.TNF-α (12.5,25,50,100,and 200 nM)was used as a positive control for the TNF-R1 sensor chip,TNF-R1(12.5,25,50,100,and 200 nM)was used as a positive control for the TNF-α sensor chip,and tetracycline(10 μM)was used as a negative control to confirm the suitability of the SPR system.The response signals were recorded and analyzed.
2.4.Recovery and identification of bioactive ingredients
The binding activity of RPA to TNF-R1 was tested by preinjection and evaluation.Gradient concentrations(0,1:1000,1:500,and 1:250)of RPA extraction diluted by the running buffer were injected into the TNF-R1 chip for 60 s,followed by disassociation for 120 s.
Then,TNF-R1-bound compounds were recovered and identified using our previously reported method[28].Briefly,the RPA sample was injected into the system,and the active ingredients were associated.A small volume recovery solution was injected to allow the bound ingredients to dissociate.Then,the recovery solution with dissolved compounds flowed back to the ammonium bicarbonate solution.These cycles were repeated five times;the recovery solutions were concentrated by nitrogen and subjected to UPLC-QTOF/MS analysis.The bioactive ingredients were identified by accurate mass data obtained by QTOF/MS and confirmed using their standard substances.
2.5.Determination of affinity constant
Three-fold serial dilutions of paeoniflorin or paeonol(0,0.14,0.40,1.23,3.70,11.1,and 33.3μM)were prepared to determine the affinity constant(KD)of paeoniflorin and paeonol towards TNF-R1.The data were analyzed using Biacore T200 evaluation software(GE Healthcare,Uppsala,Sweden),and the affinity constant was calculated.
2.6.Competition assay
Serial concentrations(0-128μM)of paeoniflorin and paeonol were first injected into the TNF-α sensor chip or TNF-R1 sensor chip for 60 s,and the response units were recorded.Then,paeoniflorin(10 or 20 μM,final concentration)or paeonol(10 or 20 μM,final concentration)was mixed with TNF-α(10 nM,final concentration)to form competitive samples and were subjected to TNF-R1 sensor chip analysis.The competitive relations between paeoniflorin and TNF-α or between paeonol and TNF-α towards TNF-R1 were evaluated by comparing the response units of different samples.
2.7.Molecular docking
The crystal structure of TNF-R1 was obtained from the Protein Data Bank(PDB ID:1EXT)[29].Water molecules and B chain were removed from the structure.The four subdomains of TNF-R1 were defined as binding sites.Molecular docking was performed using the Libdock protocol in Discovery Studio(version 3.0)and the results were visualized in PyMol.Paeoniflorin and paeonol were docked on known binding sites.Absolute energy and Libdock score were calculated,and the pose with the best absolute energy and Libdock score was considered to be the best.
2.8.Cells culture
The mouse fibroblast cell line L929 was purchased from the American Type Culture Collection(Manassas,VA,USA).Cells were cultured in a humidified incubator at 37°C in a 5% CO2atmosphere in Dulbecco's modified Eagle's medium,10% heat-inactivated fetal bovine serum,100 μg/mL penicillin,and 100 μg/mL streptomycin.
2.9.Cell viability assay
Cell Counting Kit-8(CCK8)(Dojindo,Kumamoto,Japan)assay was used to measure cell viability.Briefly,L929 cells were cultured in 96-well plates at 2×104cells/well,treated with paeoniflorin(10 and 20 μM)and paeonol(10 and 20 μM)for 3 h,and stimulated with 10 ng/mL TNF-α and 1 μg/mL Act-D for 48 h;CCK8 solution was then added according to the manufacturer's instructions.After incubation at 37°C for 1 h,the absorbance was measured using a Synergy 4 microplate reader(Biotek Instruments Inc.,Winooski,VT,USA).
2.10.Cell apoptosis assay
Apoptosis was quantified using an Annexin-V/propidium iodide(PI)double-labeled flow cytometry kit(KeyGen,Nanjing,China).L929 cells were cultured in 24-well plates at 2×105cells/well and treated with paeoniflorin(10 and 20μM)and paeonol(10 and 20 μM)for 3 h and stimulated with TNF-α and Act-D for 48 h.The cells were then collected and processed for labeling with Annexin-V-fluorescein isothiocyanate and PI.The labeled cells were analyzed using a FACSCalibur(BD Biosciences,San Diego,CA,USA).
2.11.Statistical analysis
Results are shown as mean±SD of triplicate experiments.Statistical significance was determined using Student's t-test.#P<0.05,versus the control group;*P<0.05,versus TNF-α/Act-D-stimulated group.
3.Results
3.1.Establishment and validation of screening system
Commercially available recombinant TNF-R1 protein was immobilized on a CM5 sensor chip.To demonstrate the suitability of the TNF-R1 chip,TNF-α and tetracycline were used as positive and negative controls,respectively,and were injected into the SPR system.TNF-α showed affinity toward TNF-R1,whereas tetracycline showed no binding activity(Fig.1A).This indicates that TNF-R1 immobilized on the sensor chip surface is specific to its ligands and can be applied for screening bioactive components that bind to it.
Similarly,TNF-α protein was immobilized onto the CM5 sensor surface,and the selectivity was validated using TNF-R1 and tetracycline.The results showed that only TNF-R1 could bind to the sensor chip(Fig.1B).This confirmed that the activity of TNF-α immobilized on the sensor surface was good,and the chip could be used to determine the affinity of TNF-α and the active compounds.
3.2.Screening and identification of ligands binding to TNF-R1
Then,a series of RPA extractions with different diluted ratios were injected into the sensor surface to preliminarily evaluate the interaction between potential bioactive components and TNF-R1.Interestingly,RPA extraction displayed binding activity to TNF-R1 in a concentration-dependent manner.The response signal increased with an increase in the sample concentration(Fig.2A),which suggested that some constituents in RPA extraction could bind to TNF-R1.
To reveal which ingredients could bind to TNF-R1,an injection and recovery procedure was performed using the Biacore T200 system.RPA extraction was injected into the system,and recovered samples containing TNF-R1-bound ingredients were obtained.The collected samples were concentrated in nitrogen.Identification was conducted on a UPLC-QTOF/MS system,and the total ion chromatography is shown in Fig.2B.As a result,paeoniflorin and paeonol were considered as potential active ingredients(Figs.2C and D).We then checked the extracted ion chromatography of these two compounds both in the recovered sample and RPA extraction,and found that the retention time in different samples was similar.Finally,we confirmed the retention time using paeoniflorin and paeonol standards(Figs.2E and F).This result showed that paeoniflorin and paeonol were TNF-R1-bound ingredients of RPA.
3.3.Paeoniflorin and paeonol could bind to TNF-R1 directly
To further confirm that paeoniflorin and paeonol directly interacted with TNF-R1,we determined their affinity constants by SPR assay.Serial concentrations were analyzed by SPR assay,and the affinity constants were calculated as 4.9μM for paeoniflorin(Figs.3A and B)and 11.8μM for paeonol(Figs.3C and D).The results confirmed that paeoniflorin and paeonol from RPA could bind to TNF-R1 directly.
3.4.Paeoniflorin and paeonol competed with TNF-α while binding to TNF-R1
To understand how paeoniflorin and paeonol exert their pharmacological activity,we investigated whether the interaction between these two compounds and TNF-R1 could affect the binding of TNF-α and TNF-R1.First,paeoniflorin and paeonol were tested in the TNF-α sensor chip,and no obvious response signals were observed(Figs.4A and B).This indicates that these two compounds could not interact with TNF-α but could specifically act on TNF-R1.
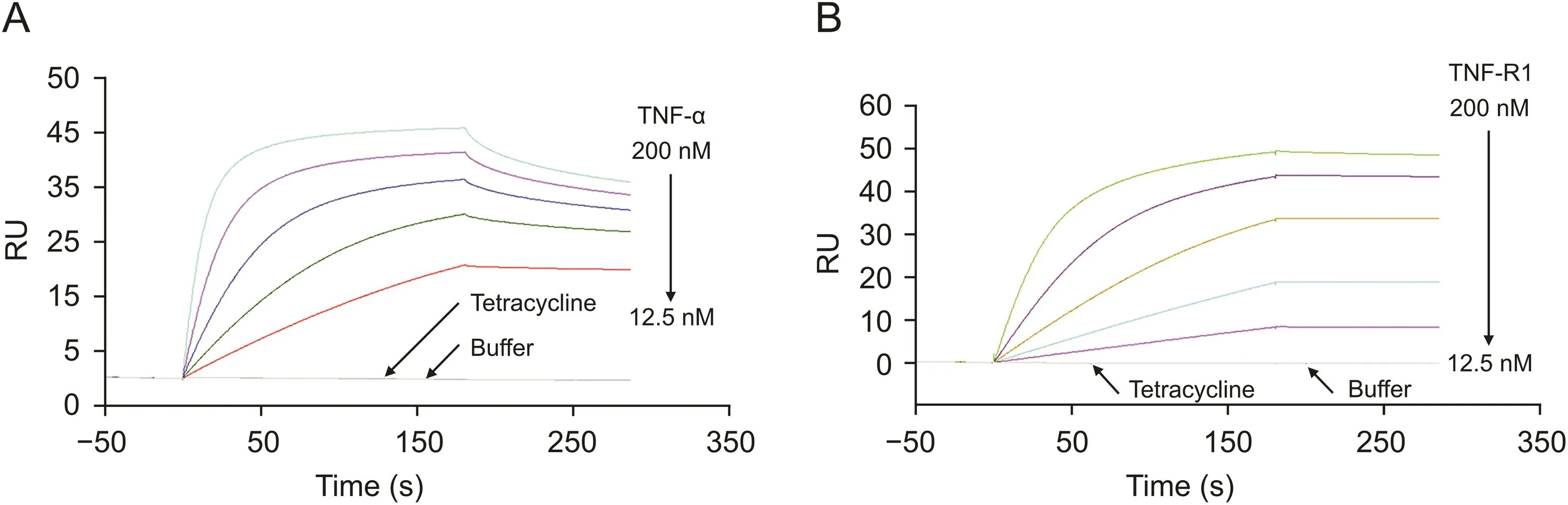
Fig.1.The bioactivity of the sensor chip was characterized by positive and negative controls after protein immobilization.(A)For tumor necrosis factor receptor type 1(TNF-R1)sensor chip,tumor necrosis factor-α (TNF-α)could bind to TNF-R1 specifically.(B)For TNF-α sensor chip,TNF-R1 could bind to TNF-α specifically.
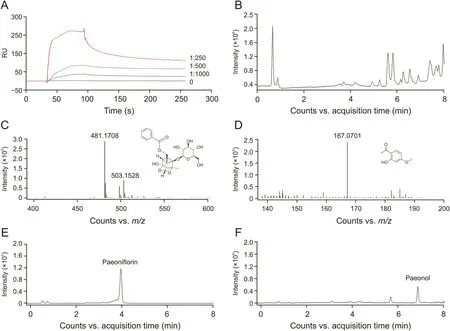
Fig.2.Recovery and identification of TNF-R1-bound ingredients from Radix Paeoniae Alba(RPA).(A)RPA extraction displayed binding activity to TNF-R1 in a dose-dependent manner.(B)Total ion chromatography(TIC)of the recovered sample.(C)Mass spectrometry and chemical structure of paeoniflorin identified from the recovered sample.(D)Mass spectrometry and chemical structure of paeonol identified from the recovered sample.(E)TIC of paeoniflorin standard.(F)TIC of paeonol standard.
Then,a competition assay was performed on the SPR system to investigate whether the two compounds compete with TNF-α while binding to TNF-R1.Paeoniflorin(10 or 20μM)was pre-mixed with TNF-α and subjected to SPR analysis.Compared to that of TNF-α alone,the response unit of the mixture decreased.In addition,the higher the compound concentration in the mixture,the lower the response(Fig.4C).This result demonstrated that paeoniflorin could compete with TNF-α while binding to TNF-R1.If they can compete with each other,a TNF-α molecule that binds to TNF-R1 will be replaced by a paeoniflorin molecule.The molecular weight of paeoniflorin(approximately 0.48 kDa)is much smaller than that of TNF-α(approximately 17 kDa);therefore,the response declines.However,if they cannot compete with each other,the response of the two molecules will increase,resulting in an increased RU of the mixture.A similar competitive relationship between paeonol and TNF-α was also observed(Fig.4D).The competition assay showed that the two compounds from RPA and TNF-α might have the same binding sites of TNF-R1 and could compete with each other;thus,paeoniflorin and paeonol will block the biological action of TNF-α on TNF-R1.
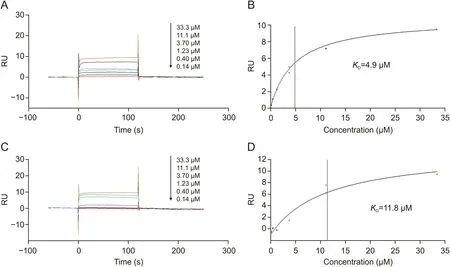
Fig.3.Affinities of the bioactive candidates towards TNF-R1 determined by surface plasmon resonance(SPR)affinity assay.(A)Sensorgrams of serial concentrations of paeoniflorin(0-33.3 μM).(B)Affinity constant of paeoniflorin.(C)Sensorgrams of serial concentrations of paeonol(0-33.3 μM).(D)Affinity constant of paeonol.
3.5.Molecular docking
Next,we performed molecular docking to predict the interplay between the two active compounds and TNF-R1.The parameters were set as follows:top hits of 100 and poster cluster radius of 0.25.Paeoniflorin and paeonol were docked on the four subdomains of TNF-R1 by Libdock.As shown in Fig.5,both paeoniflorin and paeonol had the best poses with subdomain 1 of TNF-R1.Paeoniflorin could form hydrogen bonds with Lys-75,Arg-77,and Asn-110 amino acid residues.In addition,Ser-74,Cys-76,Glu-79,Met-80,Cys-96,Leu-111,and Phe-112 amino acid residues could interact hydrophobically with paeoniflorin(Fig.5A).Paeonol could form hydrogen bonds with Arg-77,Gln-82,and Phe-112 amino acid residues.In addition,Ser-74,Lys-75,Cys-76,Asn-110,and Leu-111 amino acid residues could interact hydrophobically with paeonol(Fig.5B).The results confirmed that paeoniflorin and paeonol could effectively bind to the active site of TNF-R1.
3.6.Anti-inflammatory effects of paeoniflorin and paeonol
Inflammation is the starting point for many chronic diseases,and cell viability and apoptosis analyses are usually used to evaluate the anti-inflammatory activities of drugs.Therefore,we determined the effects of paeoniflorin and paeonol on TNF-αstimulated L929 cells.As shown in Fig.6,TNF-α and Act-D caused almost 70% cytotoxicity in L929 cells,and paeoniflorin and paeonol inhibited cytotoxicity in a dose-dependent manner.
We next explored whether paeoniflorin and paeonol can inhibit the apoptosis of TNF-α-stimulated L929 cells.As shown in Fig.7,the level of apoptosis markedly increased to 60.02% upon TNF-α and Act-D treatment,and this induction was effectively inhibited to 31.11% by 10 μM paeoniflorin or 22.31% by 20 μM paeoniflorin.Similar inhibitory effects were observed in the paeonol treatment group.This induction was inhibited to 38.3% with 10μM paeonol or 22.65% by 20μM paeonol.The results of cell viability and apoptosis suggested that the two bioactive candidates interfered with the biological activity of TNF-α and might play an important role in the anti-inflammatory activity of RPA.
4.Discussion
In this study,an effective SPR-based analysis strategy,including active ingredient screening,affinity determination,and binding site prediction,was established and used to identify and verify antiinflammatory candidates of RPA.This strategy made full use of the technical advantages of SPR and is accurate,specific,and convenient.
SPR,most commonly used to characterize biomolecular interactions,including affinity constant determination[30]and binding analysis[31,32],is considered as the most accurate way to describe the interaction between two molecules by the United States Pharmacopeia and Chinese Pharmacopeia.This technology has been increasingly used in TCM studies.For example,Ye et al.[33]studied the mechanism of Toujie Quwen granules in treating coronavirus by combining network pharmacology and molecular docking and revealed the precise protein targets of four major active components by SPR experiments.Liu et al.[34]showed that SPR could be used to compare the known α-amylase inhibitor and three citrus flavonoids binding to α-amylase and concluded that the key factor in the molecular interaction was the number and position of functional groups.
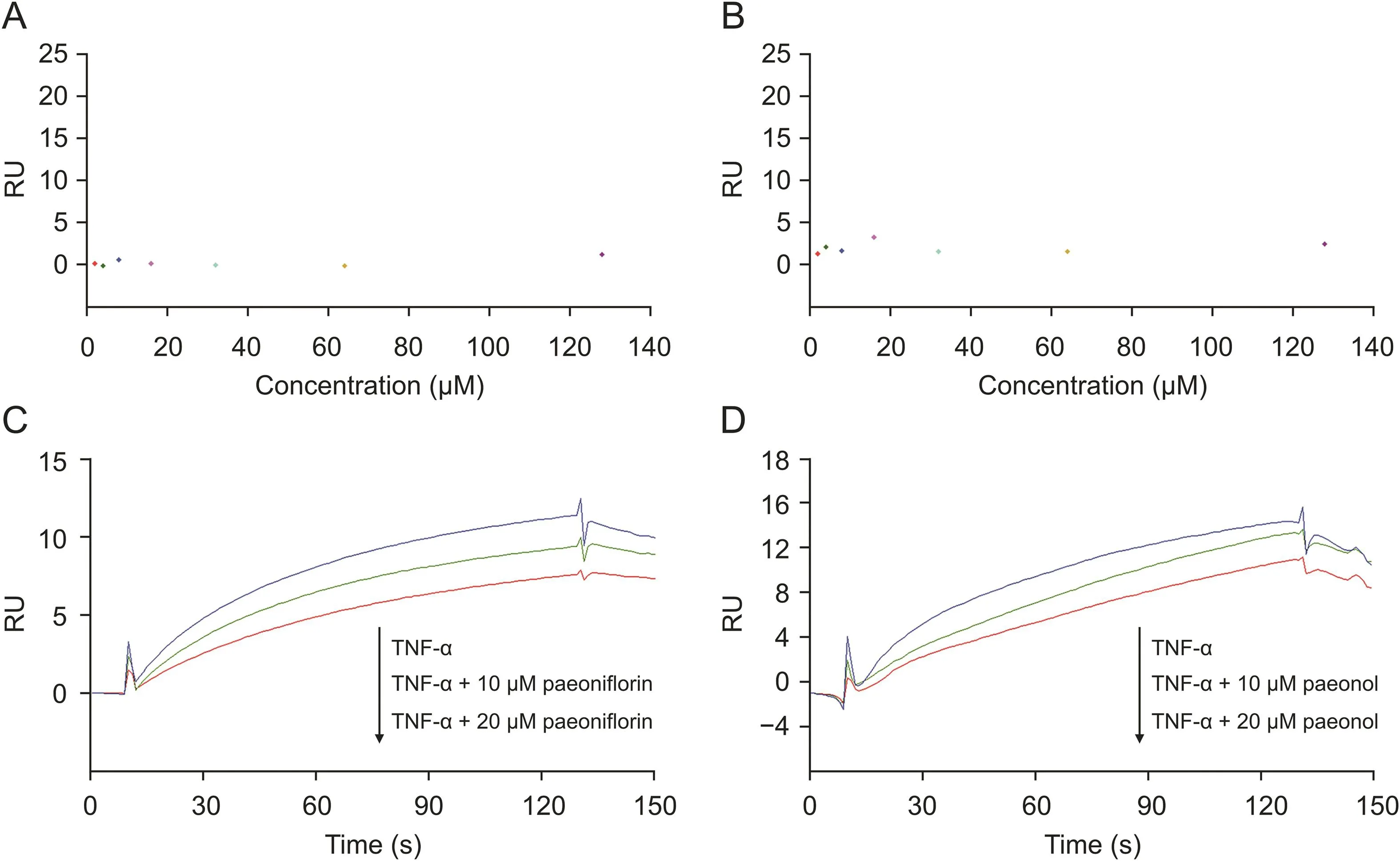
Fig.4.Competition between active candidates and TNF-α analyzed by SPR competition assay.(A)A serial concentration of paeoniflorin tested on TNF-α sensor surface showed there was no specific binding.(B)A serial concentration of paeonol tested on TNF-α sensor surface showed there was no specific binding.(C)Competition assay of paeoniflorin and TNF-α revealed that paeoniflorin could block the binding of TNF-α towards TNF-R1 in a concentration-dependent manner.(D)Competition assay of paeonol and TNF-α revealed that paeonol could block the binding of TNF-α towards TNF-R1 in a concentration-dependent manner.
Furthermore,the SPR-based active ingredient recognition system has been successfully used in the study of TCM[10-12].Interestingly,the system can specifically identify protein-bound ligands directly from compound mixtures without any prior chemical labeling.By combining affinity detection with activity screening,it is convenient to confirm the ligand-protein interaction and obtain credible screening results.This study also showed that the competition assay could be performed on the SPR system by simply mixing the molecules.The increase or decrease in the response signal of different samples reflects the competitive relationship of molecules.Competition assays are commonly used to enhance the response signal in SPR analysis,according to the principle that two molecules can bind to the same site of the target molecule.Jia et al.[35]developed an indirect competition SPR method to detect estradiol based on magnetic nanoparticles.Wei et al.[36]used SPR to detect four mycotoxins simultaneously in corn and wheat by measuring the corresponding mycotoxin antibodies after competing with free mycotoxins in samples.Thus,the SPR biosensor can not only identify the active components targeting specific proteins from TCM but also reveal the binding sites and increase understanding of their mechanism of action.Therefore,SPR technology could be applied to various aspects of drug development and,thus,has a very broad application prospect.
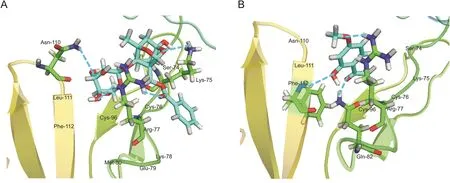
Fig.5.The interplay between TNF-R1 and(A)paeoniflorin or(B)paeonol.Paeoniflorin and paeonol had the best poses with subdomain 1 of TNF-R1.

Fig.6.Effects of paeoniflorin and paeonol on cell viability of L929 cells.L929 cells were incubated with the indicating concentrations of paeoniflorin or paeonol in the absence or presence of TNF-α and actinomycin D(Act-D)for 48 h.(A and B)Representative cell views of each group.(C and D)Cell viability was determined by CCK8 assay.Data presented are means ± SD of three independent experiments.#P<0.05 versus non-stimulated control,*P<0.05 versus TNF-α/Act-D alone by student t-test.
Sample treatment of TCM is a key step prior to the SPR-UPLCQTOF/MS screening,which ensures that the sample contains as many chemical constituents as possible.In the present study,ultrasonic extraction with 80% methanol was performed according to our method.This method has proven to be effective and compatible with SPR screening.Other methods are also available,such as traditional decoction,presoaking,microwave-assisted extraction,pressurized liquid extraction,supercritical fluid extraction,and solid-phase extraction [37-39].Sample preparation plays an important role in studying the active ingredients of TCM and affects the reliability of the screening results.Therefore,with the development of modern extraction technologies,the screening of active ingredients will be more accurate and comprehensive.
TNF-α is widely known for its proinflammatory activity and acts by interacting with two different receptors,TNF-R1 and TNF-R2[40].TNF-R1 is expressed in most cell types and therefore appears to be a more important central mediator of the TNF-α signaling pathway by recruiting key intracellular proteins to activate downstream signaling pathways.Recruitment of adaptors,including TNF receptor-associated death domain and Fasassociated death domain,can activate signaling pathways,such as apoptosis[41,42].The recruitment of other adaptor proteins,such as TNF receptor-associated factor family proteins,can lead to the activation of MAPK kinase kinase-3 and inhibitor ofκB kinase.Subsequently,the expression of the nuclear transcription factors AP-1 and NF-κB is suppressed,triggering transcriptional changes in many genes that affect cell survival and differentiation as well as immune and inflammatory responses[43,44].Paeoniflorin and paeonol were discovered as potential bioactive ingredients of RPA,and cell assays demonstrated that the two compounds could inhibit the biological functions by interfering with the interaction between TNF-α and TNF-R1 in L929 cells.Previous studies have demonstrated that paeoniflorin antagonizes TNF-α-induced apoptosis by suppressing NF-κB p65 activation[45],ameliorates atherosclerosis by inhibiting TLR4-mediated NF-κB activation[46,47],and exerts anti-psoriatic effects via the p38 MAPK pathway[48].A similar situation prevails in paeonol,which has also been reported to have various pharmacological effects,such as decreasing inflammation and immune regulation by inhibiting NF-κB activation and the MAPK signaling pathway[49].Combined with our results,it can be speculated that paeoniflorin and paeonol reduce the ability of TNFR1 to recruit adaptors by interfering with TNF-TNF-R1 interactions,thereby reducing the expressions of transcription factors,NF-κB and AP-1,and finally reducing the expressions of inflammatory mediators.These findings support the notion that the antiinflammatory activities of RPA might be due,at least in part,to paeoniflorin and paeonol,and these two compounds can be regarded as potential molecules for inflammation inhibitors.
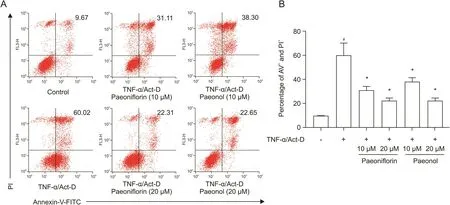
Fig.7.Effects of paeoniflorin and paeonol on apoptosis of L929 cells.L929 cells were incubated with the indicated concentrations of paeoniflorin or paeonol in the absence or presence of TNF-α and Act-D for 48 h.(A)The cells labeled with Annexin-V-fluorescein isothiocyanate(FITC)and propidium iodide(PI)were analyzed by flow cytometry.(B)The percentage of cell apoptosis was calculated.#P<0.05 versus non-stimulated control,*P<0.05 versus TNF-α/Act-D alone by Student's t-test.
In summary,this study has proved that surface plasmon resonance biosensors can be used throughout drug development,from active ingredient screening to molecular interaction analysis and binding site confirmation.By providing an example study of RPA,it is clear that SPR is a powerful tool for studying TCM.
5.Conclusions
In this study,we developed an integrated strategy that utilized SPR-based ligand screening and binding analysis and studied RPA as an example.These results support the notion that the potential bioactive components may partly account for the antiinflammatory effects of RPA.The integration of SPR-based screening,affinity determination,and binding analysis is a novel strategy to probe the bioactive ingredients of TCM and focuses on accurate biomolecular interactions covering many aspects of drug development.It can be used not only to elucidate the mechanism of TCM but also to discover novel TCM-derived drugs.
CRediT author statement
Diya Lv:Methodology,Investigation,Writing-Reviewing and Editing,Visualization;Jin Xu:Methodology,Formal analysis,Validation;Minyu Qi:Methodology,Visualization;Dongyao Wang:Validation,Resources;Weiheng Xu:Methodology;Lei Qiu:Resources;Yinghua Li:Conceptualization,Methodology,Validation,Investigation,Writing-Original draft preparation,Visualization;Yan Cao:Conceptualization,Formal analysis,Investigation,Resources,Writing-Original draft preparation,Reviewing and Editing,Supervision.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China(Grant Nos.:82003711 and 81703526)and the Shanghai Sailing Program(Grant No.:19YF1459400).
 Journal of Pharmaceutical Analysis2022年3期
Journal of Pharmaceutical Analysis2022年3期
- Journal of Pharmaceutical Analysis的其它文章
- Recent advances in quantum dots-based biosensors for antibiotics detection
- Qualitative and quantitative evaluation of Flos Puerariae by using chemical fingerprint in combination with chemometrics method
- Multiple rapid-responsive probes for hypochlorite detection based on dioxetane luminophore derivatives
- Nitrogen-doped carbon@TiO2double-shelled hollow spheres as an electrochemical sensor for simultaneous determination of dopamine and paracetamol in human serum and saliva
- Compatibility and stability studies involving polymers used in fused deposition modeling 3D printing of medicines
- A preparation strategy for protein-oriented immobilized silica magnetic beads with Spy chemistry for ligand fishing
