Association between 15 known or potential breast cancer susceptibility genes and breast cancer risks in Chinese women
Fenfen Fu,Dongjie Zhang,Li Hu,Senthil Sundaram,Dingge Ying,Ying Zhang,Shuna Fu,Juan Zhang,Lu Yao,Ye Xu,Yuntao Xie,
1Department of Breast Surgery,Peking University International Hospital,Beijing 102206,China; 2Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education),Familial & Hereditary Cancer Center,Peking University Cancer Hospital & Institute,Beijing 100142,China; 3Prenetics Limited,Hong Kong 999077,China; 4Beijing CircleDNA Gene Technology Co.,Ltd.,Beijing 100020,China
ABSTRACT Objective: There are many hereditary breast cancer patients in China,and multigene panel testing has been a new paradigm of genetic testing for these patients and their relatives.However,the magnitude of breast cancer risks related to multiple breast cancer susceptibility genes are largely unknown in Chinese women.Methods: We screened pathogenic variants in 15 established or potential breast cancer susceptibility genes from 8,067 consecutive Chinese female breast cancer patients and 13,129 Chinese cancer-free female controls.These breast cancer patients were unselected for age at diagnosis or family history.Results: We found that pathogenic variants in TP53 [odds ratio (OR): 16.9,95% confidence interval (CI): 5.2—55.2]; BRCA2 (OR: 10.4,95% CI: 7.6—14.2); BRCA1 (OR: 9.7,95% CI: 6.3—14.8); and PALB2 (OR: 5.2,95% CI: 3.0—8.8) were associated with a high risk of breast cancer.ATM,BARD1,CHEK2,and RAD51D were associated with a moderate risk of breast cancer with ORs ranging from 2-fold to 4-fold.In contrast,pathogenic variants of NBN,RAD50,BRIP1, and RAD51C were not associated with increased risk of breast cancer in Chinese women.The pathogenic variants of PTEN,CDH1, and STK11 were very rare,so they had a limited contribution to Chinese breast cancer.Patients with pathogenic variants of TP53,BRCA1,BRCA2,and PALB2 more often had earlyonset breast cancer,bilateral breast cancer,and a family history of breast cancer and/or any cancer.Conclusions: This study provided breast cancer risk assessment data for multiple genes in Chinese women,which is useful for genetic testing and clinical management of Chinese hereditary breast cancer.
KEYWORDS Multigene panel sequencing; susceptibility genes; breast cancer risk; phenotype; case-control study
Introduction
Breast cancer is the most frequent malignant tumor in Chinese women,with approximately 268,600 newly-diagnosed cases per year.Furthermore,the burden of breast cancer is still increasing1-3.It is estimated that nearly 10% of unselected breast cancer patients in China carry pathogenic variants in cancer susceptibility genes4.Detecting these pathogenic variants and precisely estimating their risks for breast cancer will provide the basis for prevention and management of hereditary breast cancers.Advances in sequencing technology have made multigene testing,or “panel testing,” a routine option to detect pathogenic variants for potential breast cancer patients and their relatives.These panels usually contain established breast cancer susceptibility genes,such asBRCA1,BRCA2,PALB2,TP53,CHEK2,andATM,and potential breast cancer susceptibility genes,such asBARD1,BRIP1,RAD50,RAD51C,andRAD51D5,6.However,the magnitude of breast cancer risks associated with these known or potential breast cancer genes is largely unknown in Chinese women.
Large case-control association studies that quantify the breast cancer risks of multiple genes from panel testing have been mainly conducted in Caucasian women or other populations7-14.However,these data may not be generally applicable to Chinese women.Although the frequencies of pathogenic variants of breast cancer genes in Chinese women are comparable to those in other ethnicities4,15,16,studies by our group or other groups have shown substantial differences in the spectrum of pathogenic variants of breast cancer susceptibility genes between Chinese and non-Chinese ethnicities,with up to one-third of the pathogenic variants in Chinese women not being found in Caucasian women17-22.Moreover,early-onset breast cancer (i.e.,diagnosed at or before the age of 40 years) is more common in Chinese women than Caucasian women1.Therefore,the breast cancer risks in specific genes in Chinese women may differ from those in other populations.In this study,we therefore screened pathogenic variants in 15 established or potential breast cancer susceptibility genes in 8,067 unselected Chinese female breast cancer patients and 13,129 Chinese cancer-free female controls,then compared the risk-related phenotypes between the patients with pathogenic variants and those without a pathogenic variant.We aimed to determine the breast cancer risks of the 15 breast cancer genes in Chinese women.
Materials and methods
Study population
A total of 8,085 consecutive breast cancer patients who were treated at the Breast Center of Peking University Cancer Hospital & Institute from October 2003 to May 2015 underwent 62-gene panel sequencing,as described in our previous report4.Eighteen male breast cancer patients were excluded from the analysis,and the remaining 8,067 female breast cancer patients were included in this study.Early-onset breast cancer patients were defined as patients diagnosed at or before the age of 40 years.Family history of breast cancer was defined as the breast cancer patient having 1 or more breast cancer patients in the first-,second-,or third-degree relatives,and family history of any cancer was defined as the breast cancer patient having 1 or more cancer patients (any kind of cancer) in the first-,second-,or third-degree relatives.The family history of breast or other cancer was collected from medical records and/or telephone interviews.A total of 13,129 Chinese women (ages ≥ 18 years) without a personal history of any cancer were recruited from the general population and were considered as a reference control for this case-control study.Written informed consents were obtained from all participants.This study was reviewed and approved by the Ethics Committee of Peking University Cancer Hospital & Institute (Approval No.2011041205) and was performed in accordance with the Declaration of Helsinki.
Sequencing assay
For the breast cancer cohort,genomic DNA extracted from peripheral blood was used for 62-gene panel sequencing at an average depth of 200-fold coverage of the target region,as described in our previous study4.We selected 15 established or potential breast cancer susceptibility genes from the 62-gene panel for further analysis based on published studies5,7,23,24,including 10 established breast cancer susceptibility genes (BRCA1,BRCA2,PALB2,TP53,PTEN,CDH1,STK11,ATM,CHEK2,andNBN) and 5 potential breast cancer susceptibility genes (BARD1,BRIP1,RAD50,RAD51C,andRAD51D).For the control cohort,genomic DNA extracted from buccal swabs was used for whole-exome sequencing at an average depth of 100-fold coverage of the target region.Reads were aligned to the reference human genome,GRCh37.Germline variations were called with GATK.Annotations were defined using ANNOVAR (https://annovar.openbioinformatics.org/en/latest/).
Variant classification
In this study,we analyzed germline variants in the 15 established or potential breast cancer susceptibility genes.Only variants with < 1% population frequency in the population databases including 1,000 Genomes (https://www.1000genomes.org),NHLBI Exome Sequencing Project (ESP6500,https://evs.gs.washington.edu/EVS/),and the Exome Aggregation Consortium (ExAC,https://exac.broadinstitute.org) were collected.Among these,the truncating variants (nonsense and frameshift variants) were included in this study,but the truncating variants in the last 55 base pairs of the penultimate exon or last exon that potentially avoided nonsense-mediated messenger RNA decay and did not influence known functional domains,were excluded.For splice-site,synonymous,nonsynonymous,in-frame,and stop-loss variants,only variants classified as pathogenic or likely pathogenic by ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) were included in the analysis.The variants with conflicting interpretations of pathogenicity in ClinVar were further annotated according to the ACMG/AMP standards and guidelines25,with supporting data from function prediction software,public literature,and curated databases (Supplementary Table S1).Variants classified to be pathogenic or likely pathogenic were considered as pathogenic in this study.
Statistical analysis
Case-control association analysis within each gene was performed using logistic regression.The strength of associations with breast cancer was estimated by the odds ratio (OR) and corresponding 95% confidence interval (CI).Genes were categorized as high risk (OR ≥ 5.0),moderate risk (2.0 ≤ OR < 5.0),or of no clinical relevance (OR < 2.0)7.Categorical variables between mutation carriers and noncarriers were compared using the chi-square test or Fisher’s exact test,where appropriate.Continuous variables were tested using at-test.Two-sidedPvalues less than 0.05 were considered to be statistically significant.All analyses were performed using SPSS 20.0 statistical software for Windows (SPSS,Chicago,IL,USA).
Results
Characteristics of the study population
The 8,067 consecutive Chinese female breast cancer patients analyzed in this study were unselected for age at diagnosis or family history.The median age at diagnosis for breast cancer patients was 50 years (range: 19—98 years) (Table 1).Among them,18.2% were diagnosed at or before the age of 40 years (early-onset breast cancer),and 10.0% had a family history of breast cancer.A total of 13,129 Chinese women without a personal history of cancer were enrolled as controls.The median age at entry was 33 years (range: 18—84 years) (Table 1).The majority of breast cancer cases (95.8%) and controls (93.6%) were of Chinese Han descent (Table 1).

Table 1 Characteristic of the study population
Pathogenic germline variants detected in breast cancer cases and controls
Pathogenic germline variants of the 15 genes were found in 654 (8.11%) breast cancer cases and 251 (1.91%) controls (Table 2).Fourteen breast cancer patients and 1 control had pathogenic variants in 2 different genes (double variant carriers; data not shown).For breast cancer cases,BRCA2(284,3.52%) was the most frequently mutated gene,followed by other established breast cancer susceptibility genes:BRCA1(146,1.81%),PALB2(57,0.71%),andTP53(31,0.38%).For controls,BRCA2(46,0.35%) still ranked as the most frequently mutated gene,followed by the candidate genes,RAD50(31,0.24%) andBRIP1(29,0.22%).Notably,pathogenic germline variants ofPTEN,CDH1,andSTK11were extremely rare in Chinese breast cancer patients or controls (Table 2).Furthermore,in all 533 pathogenic germline variants of the 15 genes analyzed in this study,385 (72.2%) variants have been reported in the latest version of the ClinVar dataset (Supplementary Table S2).
Risk estimation in the 15 genes based on this case-control study
Breast cancer risks for each of the genes were estimated by comparing the frequency of pathogenic variants identified in breast cancer cases to controls.Logistic regression results showed that 8 genes in this study were significantly associated with increased risk of breast cancer in Chinese women (Table 2).Among these genes,TP53(OR: 16.9,95% CI: 5.2—55.2);BRCA2(OR: 10.4,95% CI: 7.6—14.2);BRCA1(OR: 9.7,95% CI: 6.3—14.8); andPALB2(OR: 5.2,95% CI: 3.0—8.8) were classified as high risk breast cancer susceptibility genes in Chinese women (Table 2).BARD1(OR: 3.1,95% CI: 1.3—7.2);CHEK2(OR: 2.5,95% CI:1.4—4.6);RAD51D(OR: 2.2,95% CI: 1.3—3.8); andATM(OR: 2.1,95% CI: 1.2—3.6) were classified as moderate risk breast cancer risk genes (Table 2).The 8 high or moderate risk susceptibility genes accounted for 7.5% of unselected Chinese breast cancer patients in this study (5.2% forBRCA1/2and 2.3% for the other genes).In contrast,NBN,RAD50,BRIP1,andRAD51Cwere not associated with increased risks of breast cancer in Chinese women (Table 2).In addition,pathogenic variants ofPTEN(5 cases),CDH1(1 case),andSTK11(1 case and 1 control) were too rare to estimate their risks of breast cancer in Chinese women (Table 2).The breast cancer risks of susceptibility genes were higher after being adjusted for age (Supplementary Table S3).
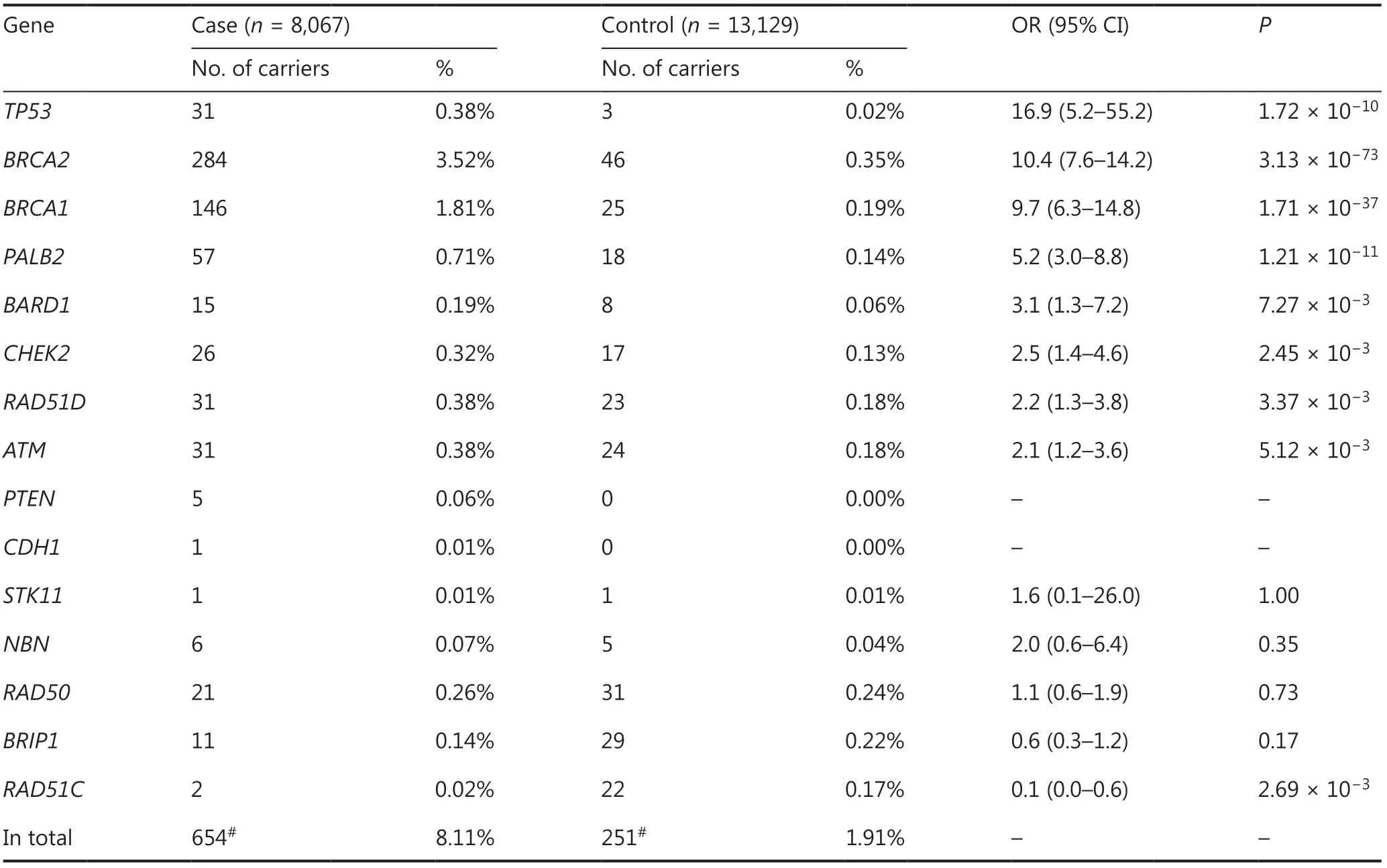
Table 2 Breast cancer risks of the 15 genes estimated by case-control association analysis in Chinese women
We further estimated breast cancer risks for each of the genes based on pathogenic truncating variants (Supplementary Table S4).Among these,BRCA1,BRCA2,andPALB2were still classified as high risk,andATM,BARD1,CHEK2,andRAD51Dwere still classifiedas moderate risk breast cancer susceptibility genes in Chinese women (Supplementary Table S4).As the majority of pathogenic variants in theTP53gene were missense,the truncating variants intheTP53gene were too rare to estimate the risk of breast cancer in this study (Supplementary Table S4).
We further analyzed the breast cancer risks of the 15 genes stratified by estrogen receptor (ER),progesterone receptor (PR),and human epidermal growth factor 2 status in this case-control study.Among the ER and/or PR+and HER2-subgroups,TP53(OR: 15.9,95% CI: 4.6—54.6);BRCA2(OR: 12.8,95% CI: 8.9—17.7);BRCA1(OR: 5.6,95% CI: 3.5—9.2); andPALB2(OR: 6.0,95% CI: 3.4—10.5) were still classified as high risk breast cancer susceptibility genes (Table 3).CHEK2(OR: 2.5,95% CI: 1.2—5.0) andATM(OR: 2.7,95% CI: 1.5—4.9) were classified as moderate risk susceptibility genes (Table 3),whileBARD1andRAD51Dwere not significantly associated with risk in this subgroup (Table 3).Among the HER2+group,onlyTP53(OR: 23.8,95% CI: 6.5—86.6) was classified as a high risk breast cancer susceptibility gene,whileBRCA2(OR: 4.5,95% CI: 2.8—7.2);BRCA1(OR: 3.4,95% CI: 1.7—6.8);PALB2(OR: 3.2,95% CI: 1.4—7.3); andCHEK2(OR: 3.8,95% CI: 1.7—8.5) were classified as moderate risk breast cancer susceptibility genes (Table 3).Among the triple negative subgroup,TP53,BRCA1,BRCA2,PALB2,BRAD1,andRAD51Dwere all classified as high risk breast cancer susceptibility genes (Table 3).Of these,BRCA1had the highest risk of breast cancer susceptibility (OR: 42.1,95% CI: 26.8—66.2) (Table 3).
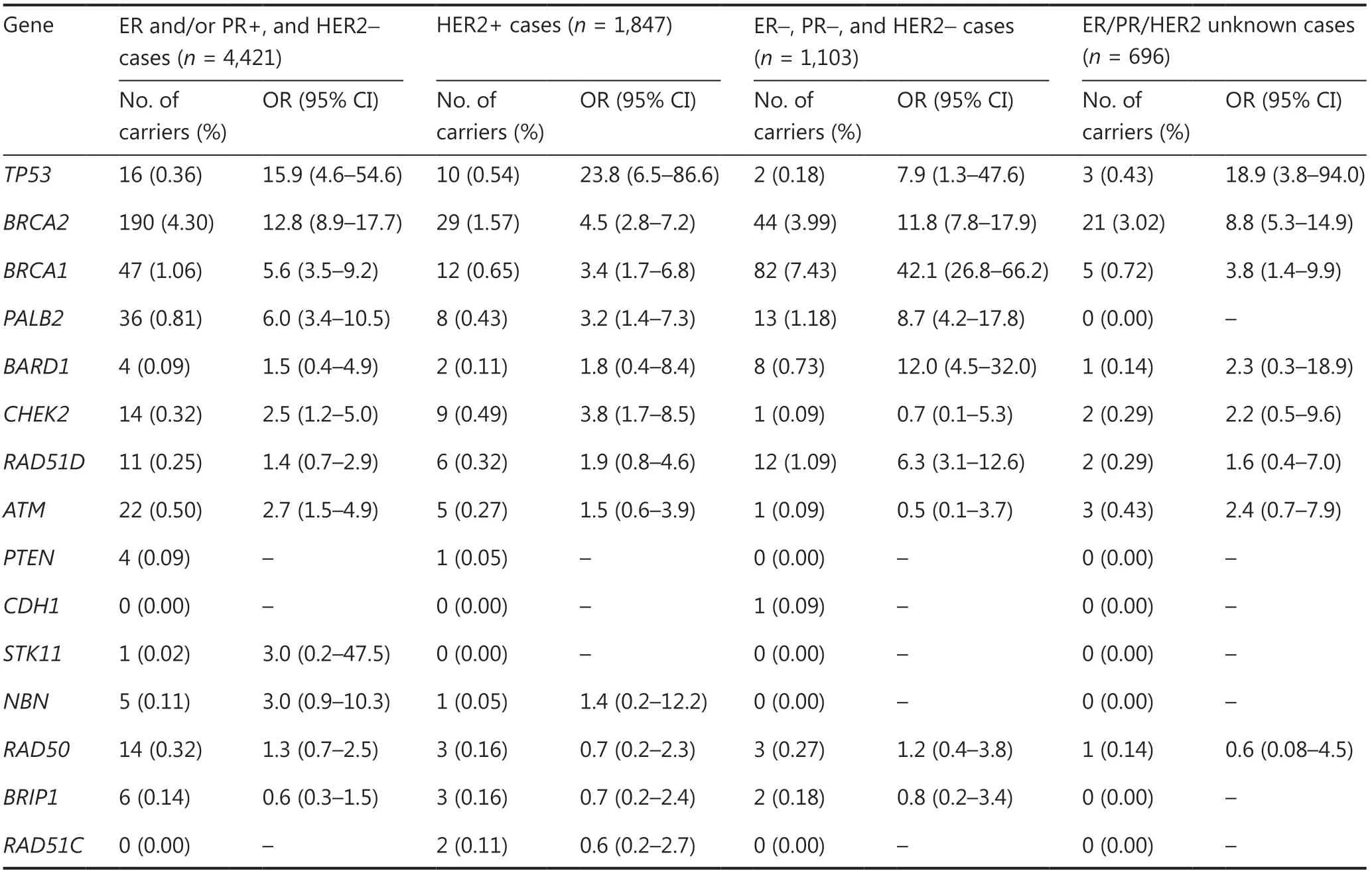
Table 3 Breast cancer risks of the 15 genes stratified by ER,PR,and HER2 status of breast cancer cases
The impact of breast cancer susceptibility genes on clinical characteristics
Pathogenic variants in breast cancer susceptibility genes are usually associated with risk-related phenotypes,such as early-onset breast cancer,bilateral breast cancer,and higher frequencies of family cancer history26.We therefore compared these risk-related characteristics between the patients with pathogenic variants and those without any pathogenic variant.Double variant carriers were excluded from the clinical characteristic analysis.
The mean age at diagnosis of breast cancer patients with pathogenic variants ofTP53(41.8 yearsvs.51.4 years,P= 5.00×10-6);BRCA2(47.8 yearsvs.51.4 years,P= 2.95×10-7);BRCA1(44.7 yearsvs.51.4 years,P= 1.01×10-11);PALB2(47.6 yearsvs.51.4 years,P= 0.02); andRAD51D(46.3 yearsvs.51.4 years,P= 0.02) was significantly younger than that of noncarriers (Table 4).Similarly,pathogenic variants ofTP53,BRCA2,BRCA1,andRAD51Dwere significantly associated with early-onset breast cancer (Table 4),and pathogenic variants ofTP53,BRCA2,BRCA1,andNBNwere significantly associated with premenopausal breast cancer (Table 4).Among these,theTP53gene,which conferred the highest risk of breast cancer,was also associated with the youngest mean age at diagnosis and the highest frequency of early-onset breast cancer (Table 4).In addition,the high risk genes (TP53,BRCA2,andBRCA1)were significantly associated with bilateral breast cancer (Table 4).When comparing the family history of cancer,all high risk and moderate risk genes except forRAD51Dwere significantly associated with higher frequencies of family history of breast cancer and/or family history of any cancer (Table 4).In summary,all 8 high or moderate risk susceptibility genes in this study affected 1 or more risk-related phenotypes.In contrast,the 4 genes that were not associated with increased risk of breast cancer in this study (NBN,RAD50,BRIP1,andRAD51C) had no effect on any of the risk-related phenotypes (Table 4).
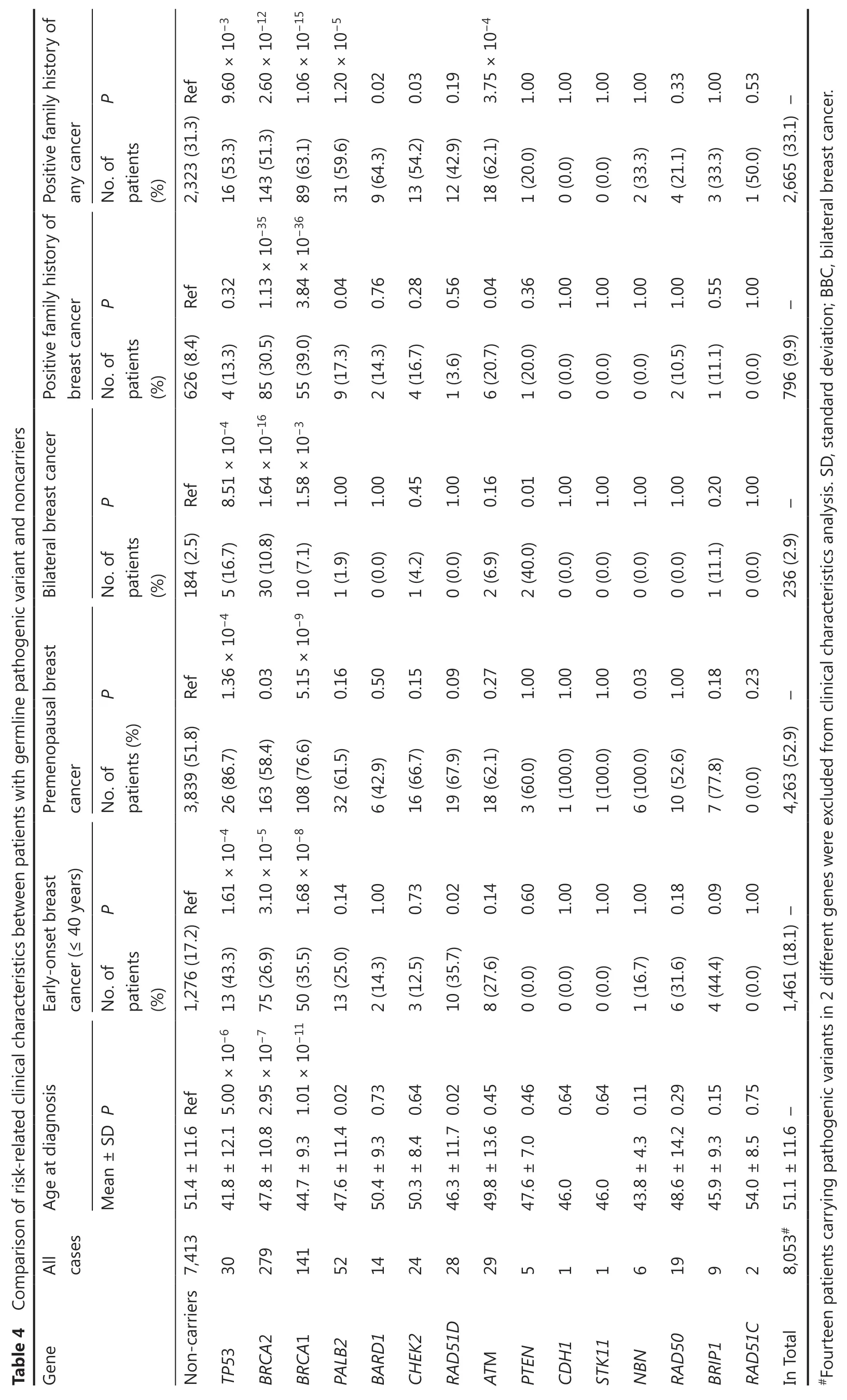
P 9.60 × 10-3 1.06 × 10-15 1.20 × 10-5 0.02 1.00 1.00 1.00 1.00 0.33 1.00 0.53 any cancer No.of patients (%) Positive family history of 2,323 (31.3) Ref 16 (53.3) 31 (59.6) 9 (64.3) 13 (54.2) 0.03 12 (42.9) 0.19 18 (62.1) 3.75 × 10-4 1 (20.0) 0 (0.0) 0 (0.0) 2 (33.3) 4 (21.1) 3 (33.3) 1 (50.0) 2,665 (33.1) - P 0.32 0.56 0.04 1.00 1.00 1.00 0.55 1.00 breast cancer No.of patients (%) 626 (8.4) Ref 9 (17.3) 0.04 2 (14.3) 0.76 4 (16.7) 0.28 1 (3.6) 6 (20.7) 1 (20.0) 0.36 0 (0.0) 0 (0.0) 0 (0.0) 2 (10.5) 1.00 1 (11.1) 0 (0.0) 796 (9.9) -Table 4 Comparison of risk-related clinical characteristics between patients with germline pathogenic variant and noncarriers P 1.00 1.00 0.45 1.00 0.16 1.00 1.00 1.00 1.00 0.20 1.00 No.of patients (%) Bilateral breast cancer Positive family history of 184 (2.5) Ref 30 (10.8) 1.64 × 10-16 85 (30.5) 1.13 × 10-35 143 (51.3) 2.60 × 10-12 1 (1.9) 0 (0.0) 1 (4.2) 0 (0.0) 2 (6.9) 2 (40.0) 0.01 0 (0.0) 0 (0.0) 0 (0.0) 0 (0.0) 1 (11.1) 0 (0.0) 236 (2.9) - P 1.36 × 10-4 5 (16.7) 8.51 × 10-4 4 (13.3) 0.03 5.15 × 10-9 10 (7.1) 1.58 × 10-3 55 (39.0) 3.84 × 10-36 89 (63.1) 0.16 0.50 0.15 0.09 0.27 1.00 1.00 1.00 0.03 1.00 0.18 0.23 cancer No.of patients (%) Premenopausal breast 3,839 (51.8) Ref 32 (61.5) 6 (42.9) 16 (66.7) 19 (67.9) 18 (62.1) 3 (60.0) 1 (100.0) 1 (100.0) 6 (100.0) 10 (52.6) 7 (77.8) 0 (0.0) 4,263 (52.9) - P 1.00 0.73 0.14 0.60 1.00 1.00 1.00 0.18 0.09 1.00 cancer (≤ 40 years)No.of patients (%) Early-onset breast 1,276 (17.2) Ref 75 (26.9) 3.10 × 10-5 163 (58.4) 13 (25.0) 0.14 2 (14.3) 3 (12.5) 10 (35.7) 0.02 8 (27.6) 0 (0.0) 0 (0.0) 0 (0.0) 1 (16.7) 6 (31.6) 4 (44.4) 0 (0.0) 1,461 (18.1) - 0.64 0.64 Mean ± SD P Age at diagnosis 41.8 ± 12.1 5.00 × 10-6 13 (43.3) 1.61 × 10-4 26 (86.7) 47.8 ± 10.8 2.95 × 10-7 44.7 ± 9.3 1.01 × 10-11 50 (35.5) 1.68 × 10-8 108 (76.6) 47.6 ± 11.4 0.02 50.4 ± 9.3 0.73 50.3 ± 8.4 0.64 46.3 ± 11.7 0.02 49.8 ± 13.6 0.45 47.6 ± 7.0 0.46 46.0 46.0 43.8 ± 4.3 0.11 48.6 ± 14.2 0.29 45.9 ± 9.3 0.15 54.0 ± 8.5 0.75 All cases 30 279 141 52 14 24 28 29 5 1 1 6 19 9 2 8,053# 51.1 ± 11.6 -Gene Non-carriers 7,413 51.4 ± 11.6 Ref TP53 BRCA2 BRCA1 PALB2 BARD1 CHEK2 RAD51D ATM PTEN CDH1 STK11 NBN RAD50 BRIP1 RAD51C In Total#Fourteen patients carrying pathogenic variants in 2 different genes were excluded from clinical characteristics analysis. SD, standard deviation; BBC,bilateral breast cancer.
TP53,PTEN,STK11,andCDH1are associated with Li-Fraumeni syndrome,Cowden syndrome,Peutz-Jeghers syndrome,and hereditary diffuse gastric cancer syndrome27-30.Among the 31 breast cancer patients withtheTP53pathogenic variant in this study,only 3 patients (9.7%) met the criteria for the Li-Fraumeni syndrome.After excluding the 3 patients with Li-Fraumeni syndrome,theTP53gene was still significantly associated with a high risk of breast cancer in Chinese women (OR: 15.2,95% CI: 4.6—50.1,P= 2.00×10-9),which suggested that women withTP53pathogenic variants but without a family history of Li-Fraumeni syndrome still had a high risk of breast cancer.None of the 5 breast cancer patients with thePTENvariant met the Cowden syndrome criteria,and the patient withSTK11variant did not meet the Peutz-Jeghers criteria.The patient with theCDH1variant in this study did not present with lobular breast cancer,nor with a personal history or family history of gastric cancer,although there were 255 (3.2%) invasive lobular breast cancer patients in this study.
Discussion
In this study,we screened pathogenic variants in 15 established or potential breast cancer susceptibility genes in 8,067 Chinese unselected female breast cancer patients and 13,129 Chinese cancer-free female controls.Our results classified
TP53,BRCA1,BRCA2,andPALB2as high risk,andATM,BARD1,CHEK2,andRAD51Das moderate risk breast cancer susceptibility genes in Chinese women.In contrast,our study revealed thatNBN,RAD50,BRIP1,andRAD51Cwere not associated with an increased risk of breast cancer,andPTEN,CDH1,andSTK11had a very limited contribution to Chinese breast cancer.
We found that theTP53gene conferred the highest risk of breast cancer in Chinese women (OR: 16.9),and patients carrying pathogenic variants of theTP53gene had the youngest mean age at diagnosis.Pathogenic variants of theTP53gene were also associated with a high risk of breast cancer in Japanese women9.However,2 recent large-scale case-control studies of Caucasian women reported that theTP53gene was not significantly associated with breast cancer risk13,14.This difference might be explained by the observation that the prevalence ofTP53pathogenic variants in Asian breast cancer patients was much higher than that in Caucasian breast cancer patients9,13,14,31.In addition,a previous study based on patients with Li-Fraumeni syndrome reported a very high relative risk of breast cancer (OR: 105) for theTP53gene27.This difference might be explained by that theTP53variant carriers in our study were identified from unselected breast cancer patients,and only a minority of them met the Li-Fraumeni Chompret criteria.Importantly,our results showed that Chinese women withTP53pathogenic variants but without a family history of Li-Fraumeni syndrome had a high risk of breast cancer (OR = 15.2).
This case-control study showed approximately a 10-fold increased risk of breast cancer forBRCA1/2pathogenic variant carriers,when compared with noncarriers in Chinese women.Recent case-control studies in Caucasian women have reported relative risks of breast cancer for theBRCA1andBRCA2genes of 5.9—10.6-fold and 3.3—5.9-fold,respectively8,13,14,which were lower than the magnitude of breast cancer risks estimated in this study.This difference might be explained by sample selection and ethnic differences.
ThePALB2gene was recently classified as a high risk breast cancer susceptibility gene in Caucasian women,with 53% cumulative risk to age 80 years and 7-fold relative risk for female breast cancer7,32,which was comparable to the breast cancer risk of theBRCA2gene in Caucasian women8,13,33.However,our results and a recent study from another group34both suggested thatPALB2conferred 5-fold increased risk for breast cancer in Chinese women,indicating that the risk associated withthePALB2gene was lower than that ofBRCA1/2genes in Chinese women,although the magnitude of risk for thePALB2gene reached the threshold of high risk genes.
This study classifiedATM,BARD1,CHEK2,andRAD51Das moderate risk breast cancer susceptibility genes in Chinese women.AlthoughATMandCHEK2were identified as classic moderate risk genes in Caucasian women35,the risks of the 2 genes in Chinese women are still worth investigating because it has been reported that the risk of pathogenic variants of the 2 genes was highly related to location and the type of variants,and considerable differences in the spectrum ofATMandCHEK2variantswere observedbetween Chinese and Caucasian ethnicities.For example,the dominant negativeATMp.Val2424Gly variant confers a much higher risk than truncating variants of theATMgene (OR: 8.0—11.0 for p.Val2424Gly and OR: 2.2 for truncating variants)35-37.CHEK2c.1100delC,the most frequent truncating variant in Caucasian women,confers a higher risk than the common missense mutation,CHEK2p.Ile157Thr38-40.However,these specific variants,such asATMp.Val2424Gly andCHEK2c.1100delC,were absent in Chinese women20,21.In summary,our study definedATMandCHEK2as moderate risk genes for breast cancer in Chinese women.Some large case-control studies in Caucasian women showed thatBRAD1andRAD51Dwere breast cancer susceptibility genes7,13,while other studies did not reach this conclusion8,14.Our results indicated thatBRAD1andRAD51Dwere moderate risk breast cancer susceptibility genes in Chinese women.In addition,breast cancer patients with theBRAD1andRAD51Dpathogenic variants were associated with a higher frequency of family cancer history and early-onset breast cancer,respectively.These clinical characteristics also suggested thatBARD1andRAD51Dwere breast cancer susceptibility genes in Chinese women.
The pathogenic variants ofPTEN,CDH1,andSTK11were too rare in our study to estimate their risks of breast cancer in Chinese women,which suggested that these genes had a very limited contribution to breast cancer in unselected Chinese women.In addition,the pathogenic variant carriers forPTEN,CDH1,andSTK11identified from unselected breast cancer patients in this study did not meet the criteria for the Cowden syndrome,hereditary diffuse gastric cancer syndrome,and Peutz-Jeghers syndrome,respectively.Additional larger case-control studies or segregation analysis in families might be needed to fully elucidate the breast cancer risks forPTEN,CDH1,andSTK11in Chinese women.NBNserves as an established breast cancer susceptibility gene in the latest guidelines,because many studies reported thatNBNc.657del5 (a founder variant in the Slavic population) was associated with increased risk of breast cancer41,42.However,NBNc.657del5 was absent in Chinese breast cancer patients and cancer-free controls in this study,and our results showed that other truncating variants ofNBNwere not associated with breast cancer risk.Our study did not supportRAD50,BRIP1andRAD51Cas breast cancer susceptibility genes in the Chinese population,consistent with results from case-control studies in Caucasian women7,8,11,14.
This study also showed that breast cancer susceptibility genes conferred different risks in breast cancer molecular subgroups.For example,pathogenic variants inBRCA1conferred the highest breast cancer risk in thetriple negative subgroup than that in other subgroups.Pathogenic variants inTP53conferred the highest breast cancer risk in theHER2+subgroup than that in other subgroups.In addition,theATMandCHEK2genes were classified as moderate risk susceptibility genes in the ER and/or PR+and HER2-subgroups,but were not associated with a risk in the triple negative subgroup.These findings were consistent with previous case-control studies8,12-14.
This study had some limitations.The ages of the cancer-free controls were younger than those of the breast cancer cases.It therefore may have underestimated the risks based on logistic regression models.However,our previous study based on the kin-cohort method found that the cumulative breast cancer risks inBRCA1/2carriers up to the age of 70 years were 37.9% and 36.5% in Chinese women,respectively43,which were also 10-fold higher risks than noncarriers (3.6% by the age of 70 years).Similar results based on 2 different methods confirmed the reliability of our findings,although some bias existed in this case-control study.
Conclusions
This large case-control study classifiedTP53,BRCA2,BRCA1,andPALB2as high risk,andATM,BARD1,CHEK2,andRAD51Das moderate risk breast cancer susceptibility genes in Chinese women.In contrast,the study showed thatNBN,RAD50,BRIP1,andRAD51Cwere not associated with an increased risk of breast cancer,andPTEN,CDH1,andSTK11had a very limited contribution to Chinese breast cancer.These results suggested that the 8 high or moderate risk susceptibility genes should be covered in panel testing for high risk Chinese breast cancer patients and their relatives.Overall,our risk assessment data should be useful for the prevention and early detection of Chinese women who carry a pathogenic variant in the 8 breast cancer susceptibility genes.
Grant support
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos.81772824,81372832,and 81974422).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Conceived and designed the analysis: Yuntao Xie and Li Hu.
Collected the data: Fenfen Fu and Dongjie Zhang.
Contributed data or analysis tools: Senthil Sundaram,Dingge Ying,Ying Zhang,Shuna Fu,Juan Zhang,Lu Yao,and Ye Xu.
Performed the analysis: Fenfen Fu,Dongjie Zhang,and Li Hu.
Wrote the manuscript: Li Hu and Yuntao Xie.
 Cancer Biology & Medicine2022年2期
Cancer Biology & Medicine2022年2期
- Cancer Biology & Medicine的其它文章
- Erratum to A truncated protein product of the germline variant of the DUOX2 gene leads to adenomatous polyposis
- Highlighted multi-modifications of enzymes: a novel succinylation mediated by histone acetyltransferase 1 in tumors
- Systematic screening reveals synergistic interactions that overcome MAPK inhibitor resistance in cancer cells
- Efficacy of rigosertib,a small molecular RAS signaling disrupter for the treatment of KRAS-mutant colorectal cancer
- Updates in endocrine therapy for metastatic breast cancer
- The advance of adjuvant treatment for triple-negative breast cancer
