OstMAPKKK5,a truncated mitogen-activated protein kinase kinase kinase 5,positively regulates plant height and yield in rice
Yhui Liu,Yu Zhu,Xuding Xu,Fn Sun,Jingshui Yng,Liming Co*,Xiojin Luo,c,**
aState Key Laboratory of Genetic Engineering, Institute of Genetics,School of Life Sciences,Fudan University,Shanghai 200438,China
bInstitute of Crop Breeding and Cultivation, Shanghai Academy of Agricultural Sciences,Shanghai 201403,China
cEngineering Research Center of Gene Technology,Ministry of Education,Fudan University,Shanghai 200438,China
Keywords:OstMAPKKK5 Plant height and yield Cell size Gibberellin Oryza sativa Available online 6 April 2019
ABSTRACT Rice (Oryza sativa L.) is a major food crop worldwide. Plant height and yield are important agronomic traits of rice. Several genes regulating plant height and/or yield have been cloned. However, the molecular mechanisms coordinating plant height and yield are not fully characterized. Here, we report a novel gene, OstMAPKKK5 that encodes a truncated variant of a mitogen-activated protein kinase kinase kinase 5 (OsMAPKKK5) lacking an intact kinase domain.Transgenic plants overexpressing OstMAPKKK5 in indica cultivar 9311 showed increased plant height,grain length,grain width,1000-grain weight,grain number per main panicle, and yield. Real-time quantitative PCR showed that OstMAPKKK5 was widely expressed in various tissues and developmental stages. The increased plant height and yield were attributed to enlarged cell size.Overexpression of OstMAPKKK5 led to higher contents of various forms of endogenous gibberellin (GA), especially the most common active forms, GA1, GA3, GA4. We concluded that OstMAPKKK5 positively regulates plant height and yield in rice by affecting cell size,and that its underlying mechanism is based on increased endogenous GA content.
1.Introduction
Rice(Oryza sativa L.)is a major food crop,and key plant model for monocot research due to its small genome size (~400 Mb)[1]. Plant height and yield are important agronomic traits for cereal production [2]. Plant hormones gibberellins (GAs) and brassinosteroids (BRs) play major roles in modulating plant height [3]. Individual plant yield is a complicated agronomic trait determined by three component traits: number of panicles per plant, number of grains per panicle, and grain weight, among which, the most dependable determinant is grain weight.Grain shape is characterized by a combination of grain length, grain width, grain length-to-width ratio, and grain thickness, each of which is positively correlated with grain weight [2,4]. Many genes controlling either plant height or plant yield have been cloned[5-14].For example,OsMAPK6 influences rice grain size and biomass production [15,16];DTH8/Ghd8 regulates plant height and yield potential, while suppressing flowering [17,18]; and the Short Grain Length (SGL)gene, which encodes a kinesin-like protein with transactivation activity, are involved in regulation of grain length and plant height in rice [19]. However, the integration of underlying mechanisms is not yet well characterized.
Mitogen-activated protein kinase(MAPK)cascades made up of mitogen-activated protein kinase kinase kinase (MAPKKK),mitogen-activated protein kinase kinase (MAPKK) and mitogen-activated protein kinase(MAPK),are conserved signal transduction modules in eukaryotes, and are associated with various biological processes such as cell division, hormone response, plant development, disease resistance, and abiotic stress [20,21]. For example, the YODA-MKK4/5-MAPK3/6 module plays a critical role in stomatal patterning and inflorescence architecture [22,23] and the ANP-MKK6-MAPK4 cascade is required for cytokinesis [24,25]. The OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice, positively regulates grain size and weight, whereas OsMKP1 represses grain growth by directly interacting with,and deactivating,OsMAPK6[15,16,26].These studies imply that plants may have adopted different combinations of MAPKKK-MAPKK-MAPK proteins to regulate different biological processes. As the first level of this phosphorylation cascade, the MAPKKK family has the largest number of members, along with more complex and variable primary structures and domain compositions[27].The roles of individual plant MAPKKK proteins have been reported. The NPK1 gene from tobacco (Nicotiana tobacum) was shown to regulate cell division [28-30], and overexpression of its kinase domain (KD) in maize (Zea mays) enhanced abiotic stress tolerance [31]. ANP1, the homolog of NPK1 in Arabidopsis thaliana, is responsive to oxidative stress and involved in negative regulation of auxin signaling [32,33]. In addition,both CTR1 that negatively regulates ethylene response [34],and EDR1 that is involved in powdery mildew resistance and salicylic acid-inducible defense response in Arabidopsis[35,36],are MAPKKK family proteins. Using whole-genome phosphorylation analysis Tang et al. [37] found that MAPKKK5 was a potential substrate of BSK1 in Arabidopsis.BSK1 interacts with,and phosphorylates,MAPKKK5 to regulate immunity to bacterial or fungal pathogens in Arabidopsis.Raf-like MAPKKK genes DSM1 and ILA1 were thoroughly studied in rice.DSM1 mediated drought resistance through reactive oxygen species scavenging[38] and ILA1 interacted with a nuclear protein family, and regulated mechanical tissue formation in lamina joints [39].However,the role of MAPKKK in regulating height and yield in rice remains largely unknown.
In this study we identified a novel gene that encodes a naturally truncated mitogen-activated protein kinase kinase kinase 5 (OsMAPKKK5), and named it OstMAPKKK5. Expression of OstMAPKKK5 occurred in many tissues and at various growth stages, but decreased with approaching maturity. Overexpression of OstMAPKKK5 in rice cultivar (cv.) 9311 led to increased plant height and yield.Paraffin sectioning and scanning electron microscopy showed that the increased plant height and yield were caused by larger cell size. The levels of various forms of gibberellin were higher in transgenic plants overexpressing OstMAPKKK5. We concluded that OstMAPKKK5 positively regulates plant height and yield in rice as a consequence of increased endogenous gibberellin levels.
2. Materials and methods
2.1. Plant materials and growth conditions
Subspecies indica cv. 9311 was used in this study. Transgenic plants overexpressing OstMAPKKK5 were identified and used in further analyses.All plants used in the study were grown in fields of the experimental stations in Shanghai (31°11′N,121°29′E) and Sanya (18°14′N, 109°31′E) in Hainan province,or in a greenhouse at 28 °C with a 14 h light/10 h darkness photoperiod at Fudan University in Shanghai.
2.2. Microarray
Two independent biological replicates of spikelets of wild type cv. 9311 and transgenic plants overexpressing FCA-RRM2 at the booting stage under field conditions were used for microarray experiments. Microarray experiments were performed following the standard protocols for Affymetrix GeneChip services(CapitalBio,Beijing).
2.3. Bioinformatic analyses
The sequences of full-length cDNAs of OsMAPKKK5 and OstMAPKKK5 were obtained from the National Center for Biotechnology Information, U.S. National Library of Medicine(https://www.ncbi.nlm.nih.gov/gene/) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). Domain predictions for OsMAPKKK5 and OstMAPKKK5 were made using the SMART database (http://smart.emblheidelberg.de/).
2.4. Plasmid construction and rice transformation
A 1.35 kb coding sequence of OstMAPKKK5 amplified from the cDNA library of wild type 9311 was cloned into the Nco I and BstE II sites of pCAMBIA 1304 to make a pCAMBIA1304-35S::OstMAPKKK5 construct that introduced into cv.9311 callus by microprojectile bombardment using a gene gun to generate overexpression of OstMAPKKK5 transgenic lines.
2.5. Cellular analysis
The uppermost internodes and spikelets of OstMAPKKK5 overexpressing transgenic plants and wild type cv.9311 were collected at the booting stage. The internodes were fixed in formalin-acetic acid-alcohol (50% ethanol, 5% glacial acetic acid and 3.7% formaldehyde) overnight at 4 °C and then dehydrated in a graded alcohol series. Samples were embedded in Paraplast (Sigma-Aldrich, http://www.sigmaaldrich.com/) and sliced. Sections stained with 0.5% Fast Green were observed using a light microscope(Leica,Germany).Spikelets were fixed with isopentyl acetate, dried in a critical point drier, gold sputter-coated, and observed under a scanning electron microscope(Hitachi TM3000,Japan).
2.6. RNA isolation and quantitative RT-PCR
Total RNAs were isolated from various tissues using a MiniBEST Universal RNA Extraction Kit (TaKaRa, Otsu,Japan). First-strand cDNAs were synthesized with a PrimeScript RT reagent kit (TaKaRa) and used for PCR. Realtime quantitative RT-PCR (qPCR) experiments were conducted on a Bio-Rad CFX96 real-time PCR system (Bio-Rad,Hercules,CA,USA)using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). For semi-quantitative RT-PCR, amplifications were performed in 20 μL volumes using the following protocol: one cycle of 95 °C for 30 s, 35 to 40 cycles of 95 °C for 15 s,60 °C for 30 s.Rice Actin gene(OsActin1)was used as a gene reference for normalizing transcript levels. The relative quantification method (ΔΔCt) was used to evaluate quantitative variation of replicates examined. Primers are listed in Table S1.
2.7. Hormone measurements
For transgenic lines overexpressing OstMAPKKK5, about 50 germinated seedlings of each line were grown in a row in a metal box(80 cm × 55 cm × 8 cm)along with the wild type cv.9311 control.Leaves from 40 3-leaf stage seedlings of each line were harvested, quick-frozen and ground in liquid nitrogen.Measurements were made on each sample with three technical replicates.Levels of endogenous GA were measured using the derivatization approach coupled with nano-LC-ESIQ-TOF-MS analysis as described[40].
3. Results
3.1. Identification of OstMAPKKK5 and overexpression ofOstMAPKKK5 in rice
Flowering control locus A(FCA)protein,a nuclear protein and strong promoter of transition from the vegetative state to flowering in Arabidopsis has two RNA-binding motifs (RRMs)[41].Many reports on FCA-RRMs were related to their roles in floral development [42]. Previously, we found that yield and cell size in rice were increased by overexpression of RRMs FCARRM1 and FCA-RRM2 [43,44]. Yield and cell size in cotton(Gossypium hirsutum) and oilseed rape (Brassica napus) were increased by overexpression of B. napus FCA-RRM2 [45,46].Subsequent microarray analysis of spikelets from FCA-RRM2 overexpression transgenic plants at the booting stage identified a novel transcript of OsMAPKKK5,named OstMAPKKK5(GenBank:AK106496.1), that was 25-fold upregulated compared with the wild type, whereas expression of the normal OsMAPKKK5(GenBank: AK068541.1) transcript showed no significant difference. Unlike the normal OsMAPKKK5 transcript, OstMAPKKK5 retains the second intron that causes premature termination of translation. The consequence is that OstMAPKKK5 encodes a protein lacking an intact serine/threonine protein kinase,catalytic domain(S_TKc domain)(Fig.1).
To investigate whether the highly upregulated expression of OstMAPKKK5 was associated with increased grain size and weight, we transformed the OstMAPKKK5 expression vector into cv. 9311 by particle bombardment. Expression levels of OstMAPKKK5 in transgenic plants were examined in two T1transgenic plants(MD01 and MD211)and in wild type cv.9311 plants. Expression of OstMAPKKK5 in MD01 and MD211 was 1.35-fold and 4.33-fold greater than in cv. 9311, respectively(Fig. 2-B). These results demonstrated that OstMAPKKK5 was strongly expressed and genetically transmitted to the T1generation. Seeds of the T1lines were collected for further experiments.
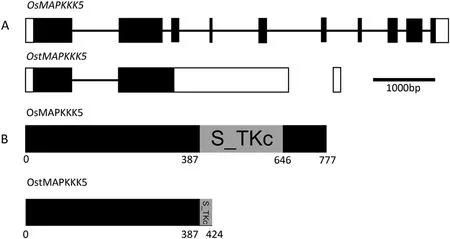
Fig.1- Schematics of OsMAPKKK5 and OstMAPKKK5 and the proteins they encode.(A)Gene structures of OsMAPKKK5 and OstMAPKKK5.Untranslated regions and exons are indicated with white and black boxes,respectively.Introns are indicated by lines.Bar,1000 bp.(B)Domain structures of OsMAPKKK5 and OstMAPKKK5.Numbers represent amino acid sequence numbers.Grey boxes indicate the S_TKc domain(serine/threonine protein kinase,catalytic domain)in OsMAPKKK5,which is terminated in OstMAPKKK5.

Fig.2- Phenotypic characterization of cv.9311 and transgenic plants overexpressing OstMAPKKK5.(A)Morphologies of cv.9311(left)and transgenic plants MD01(middle)and MD211(right).Bar,18 cm.(B)Expression levels of OstMAPKKK5 determined by qPCR in cv.9311 and transgenic plants.(C)Internode lengths of cv.9311 and transgenics.P,panicle;I to VI indicate the first to the sixth internode from the top, respectively(n = 10 for each line). (D)Comparison of grain lengths of cv.9311(top)and transgenics.Bar,1 cm.(E-I)Analyses of grain length(E),grain width(F),grain number per main panicle(G),1000-grain weight(H),and yield per plant(I)(n = 12 for each line).Values in(B,C,E-I)are means±SD.*P <0.05,**P <0.01,comparisons with the wild type using Student's t-tests.
3.2. Overexpression of OstMAPKKK5 in rice increases plant height and yield
The phenotypes of transgenic plants MD01 and MD211 were examined to understand the functions of OstMAPKKK5. Plant height and yield were significantly increased (Fig. 2-A). The increased plant height relative to cv. 9311 was largely due to elongation of the panicle and the first second internodes(Fig.2-C).All yield-related traits of the transgenic lines,including grain number per main panicle(Fig.2-G),1000-grain weight(Fig.2-H),and yield per plant (Fig. 2-I) were significantly increased compared with the wild type. Grain length and width (Fig.2-DF)in the transgenic plants were also significantly larger than in the wild type. These results showed that overexpression of OstMAPKKK5 promoted plant height and yield.
3.3.Enlarged cell size caused increased plant height and larger grain size in lines overexpressing OstMAPKKK5
To understand the function of OstMAPKKK5 at the cellular level, we used paraffin sectioning and scanning electron microscopy (SEM) to compare the two overexpression lines with cv. 9311. As the increase in plant height came from increases in the lengths of uppermost internodes, we sampled tissues from the first internode (Fig. 3-A). The cells were significantly longer and wider than those in cv.9311(Fig.3-B,C).In addition,cells from mid sections of lemmas(Fig.3-D)were also significantly larger in the transgenic lines.As shown in the scanning electron microscope images (Fig. 3-E, F), the inner epidermal cells of lemmas from OstMAPKKK5 transgenic plants were significantly larger than those from cv.9311(Fig.3-G, H). These results indicated that enlarged cell size in transgenic lines overexpressing OstMAPKKK5 led to increased plant height and large grain size.
3.4. Expression pattern of OstMAPKKK5
For determination of spatial and temporal expression of OstMAPKKK5, we performed qPCR analyses (Fig. 4) of tissues sampled at the three-leaf,tillering and booting growth stages.Expression of OstMAPKKK5 was apparent in all the tissues examined.Expression of OstMAPKKK5 appeared to decrease as plants entered the flowering stage. Expression was much higher in leaves than in other tissues. These results were consistent with the possible function of OstMAPKKK5 in regulation of plant height and yield, as both traits are largely determined prior to flowering.

Fig.3- Cellular analysis of cv.9311 and transgenic lines overexpressing OstMAPKKK5(OE#OstMAPKKK5). (A)Longitudinal sections of the uppermost internode.Bar,200 μm.(B-C)Cell length(B)and cell width(C)comparisons in paraffin sections(n =100).(D)Seed morphologies showing regions used for cytological examination.Bar,5 mm.(E-F)Scanning electron microscopy analyses of the outer(E) and inner(F)surfaces of lemmas. Bar,100 μm(E),90 μm(F).(G-H)Cell lengths(G) and widths(H)observed under scanning electron microscopy(n = 40 for each line).Values in(B, C,G,and H)are means±SD.* P <0.05,**P <0.01 compared with cv.9311 using Student's t-test.

Fig.4- Expression profile of OstMAPKKK5 in wild type 9311 indica rice. Each sample comprised a pool of 10 individuals and measures were made three times.Transcript levels were normalized using OsActin1 as a reference gene.
3.5.Overexpression of OstMAPKKK5 causes increased gibberellin contents
As gibberellins (GA) are key regulators of plant height and yield, we hypothesized that the increased plant height and yield in plants overexpressing OstMAPKKK5 was related to increased endogenous GA levels. To test this hypothesis, we measured the amounts of endogenous GA in T2generation transgenic plants and cv. 9311. The transgenic plants had higher GA contents than cv. 9311 (Fig. 5). Three commonly active forms,GA1,GA3,and GA4,showed the most significant increases in the transgenic plants compared with the wild type cv.9311.Notably,both MD01 and MD211 produced GA4,whereas cv. 9311 did not. Other GA forms showed 1.64- to 9.40-fold higher levels than cv. 9311. These results indicated that overexpression of OstMAPKKK5 led to increased GA contents, which in turn caused the increased plant height and yield in lines overexpressing OstMAPKKK5.
4. Discussion

Fig.5-Contents of various forms of gibberellin(GA)in wild type and transgenic plants overexpressing OstMAPKKK5,MD01 and MD211,respectively.Each sample was measured with three technical replicates.
We identified a novel gene, OstMAPKKK5, regulating plant height and yield in rice. A comparison of internode length among transgenic plants and wild type cv. 9311 showed that the lengths of the first and second internodes and panicles of the transgenic plants were significantly longer than those of cv. 9311, but there were no significant differences between the lengths of the other internodes(Fig. 2-C). Yield traits of the transgenic plants, including grain length, grain width, grain number on the main panicle, 1000-grain weight and yield per plant, were consistently higher than cv. 9311 (Fig. 2-A, D-I). Several genes have similar functions to OstMAPKKK5, such as OsMAPK6, Ghd8, DTH8 [15,17,18]. Many genes influence plant height and yield by regulating cell proliferation and/or cell expansion [47], such as OsSPL13, OsMAPK6, SGL[15,19,48]. Paraffin sectioning and scanning electron microscopy showed that the increased plant height and yield are a result of enlarged cell size in leaves,stems and glumes(Fig.3). Thus,OstMAPKKK5 mediates plant height and grain size by controlling cell size.
Expression of OstMAPKKK5 was detected at all growth stage and in various tissues, and was most abundant in the leaves(Fig. 4). These results provide further evidence that OstMAPKKK5 functions in regulating plant height and yieldrelated traits. The increases in plant height and yield caused by the overexpression of OstMAPKKK5 were associated with increased GA levels(Fig.5).We propose that overexpression of OstMAPKKK5 caused the increase in GA content, in turn leading to enlarged cell size, and ultimately to increased plant height and yield.
Previously, we found that overexpression of RRMs FCARRM1 and FCA-RRM2 led to increased cell size and yield in rice[43,44]. Overexpression of B. napus FCA-RRM2 similarly resulted in increased yield and cell size in cotton (Gossypium hirsutum) and oilseed rape (Brassica napus) [45,46]. These results suggest that FCA-RRM1 and FCA-RRM2 have roles in regulation of cell size. Microarray results of transgenic plants overexpressing FCA-RRM2 indicated upregulated expression of OstMAPKKK5. The present study provides evidence that OstMAPKKK5 regulates GA levles and cell size, thereby increasing plant height and yield. Further study is needed for a better understanding of the signaling pathway involving RRMs and OstMAPKKK5 and their influence on GA content.
In summary, we demonstrated that OstMAPKKK5, a truncated MAPKKK, has a role in regulating GA levels and thereby regulates cell size that in turn affects both plant height and yield. These results contribute to a better understanding of the molecular mechanisms that underlies plant height and yield.
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2019.03.001.
Acknowledgments
We thank Dr. Linhan Sun for the help and guidance on manuscript preparation. This work was supported by the National Natural Science Foundation of China (31471461,31671655), the National Transgenic Major Project of China(2016ZX08001004-001), and Shanghai Agriculture Applied Technology Development Program,China(G2014070102).
- The Crop Journal的其它文章
- Suppression of starch synthase I (SSI) by RNA interference alters starch biosynthesis and amylopectin chain distribution in rice plants subjected to high temperature
- Fine mapping of qTGW10-20.8,a QTL having important contribution to grain weight variation in rice
- Characterization of a new hexaploid triticale 6D(6A)substitution line with increased grain weight and decreased spikelet number
- TaCML36,a wheat calmodulin-like protein,positively participates in an immune response to Rhizoctonia cerealis
- Ameliorative effects of potassium on droughtinduced decreases in fiber length of cotton(Gossypium hirsutum L.) are associated with osmolyte dynamics during fiber development
- Exogenous application of glycine betaine improved water use efficiency in winter wheat(Triticum aestivum L.) via modulating photosynthetic efficiency and antioxidative capacity under conventional and limited irrigation conditions