Prevalence and genetic divergence of Leptospira interrogans and Leptospira borgpetersenii in house rats (Rattus rattus) from Peninsular Malaysia
Nurul Huda Mohamad Ikbal, Subha Bhassu, Khanom Simarani, Shigehiko Uni, Chew Chin Chan,Hasmahzaiti Omar
Institute of Biological Sciences, Faculty of Science, University of Malaya, 50603 Kuala Lumpur, Malaysia
ABSTRACT Objective: To evaluate the prevalence and divergence of genetically identified Leptospira spp.in the population of Rattus rattus.Methods: A total of 130 rats were used in this study. The infection within the rats were screened using polymerase chain reaction (PCR)-based diagnosis, with Leptospira genusspecific 16S rRNA primer and pathogenic Leptospira spp. specific LipL32 primer, on both kidney and liver tissues of Rattus rattus to detect the presence of potential Leptospira spp.Results: Out of 130 rats studied, 51 (39.23%) individuals were positive for leptospiral DNA.Basic Local Alignment Search Tool (BLAST) and phylogenetic analysis revealed that both pathogenic Leptospira interrogans and Leptospira borgpetersenii were predominantly identified.Phylogenetically, both genes disclosed similar clustering patterns of tree topologies between the two species. Although both genes were conserved, LipL32 gene portrayed higher nucleotide divergence (5.80%) compared to the 16S rRNA gene (0.60%). Minimum-spanning network displayed several haplotypes that are unique to each species, suggesting a higher degree of subdivision between both species. As for prevalence surveillance, both adult and subadult rats were susceptible to the infection, in which males were the most susceptible. Kidney was notable as the favourable organ for colonisation of leptospires. Rats captured from fresh markets were highly infected with Leptospira spp. (54.28%) compared to those from housing areas (26.47%).Conclusions: Rattus rattus represents an important asymptomatic transmitter of pathogenic leptospires, and hence is of public health concerns.
Keywords:Pathogenic leptospires Public health PCR Molecular diagnostic Zoonotic
1. Introduction
Leptospirosis is one of the most prevalent zoonotic and emerging infectious diseases promoted by the pathogenic spirochete genus Leptospira[1]. At least 22 genomic Leptospira spp. were classified via DNA-DNA hybridisation analysis and over 300 serovars were identified from agglutinating lipopolysaccharide (LPS) antigens[2].These species can be categorised into pathogenic, intermediate, and saprophytic species[2,3]. The detailed species and their belonging categories were well-elaborated in previous works[2,4].
Leptospirosis is transmitted directly or indirectly through the urine of infected reservoir animals[5,6]. Rodents are presumably the main reservoirs of Leptospira spp. among other wild and domestic animals [7]. Rattus(R.) rattus (black rat) is a species of rodents globally distributed that is often related to leptospiral infection[8]. These rats are currently appraised as the primary maintenance hosts of Leptospira spp. and the conveyors for pathogenic leptospiral serovars[9].
Prior research on leptospirosis in Malaysia concentrated mostly on humans and domestic animals in the outbreak areas[10,11]. The rat's population in Malaysia is increasing at an alarming rate, yet there is still insufficient documentation regarding this infection on urban rat populations in the region. Recently, two predominant pathogenic leptospiral serovars were seen, Leptospira (L.) borgpetersenii serovar Javanica and L. interrogans serovar Bataviae, dispersed by two dominant rat species, R. rattus and R. norvegicus in Peninsular Malaysia[12]. An additional of two Leptospira spp. of L. noguchii and L. weilli were then discovered in wild rats from Kuala Lumpur[13].
Lately, molecular approaches such as conventional and realtime PCR are acknowledged as the fastest validation methods for early-phase infection of the disease and negate the requirement for isolation and culture of the disease-causing organism. These procedures can be conducted reliably on numerous templates,involving blood, kidney tissues, and urine[14]. For screening of infection, PCR using Leptospira genus-specific primer of 16S rRNA gene and pathogenic Leptospira spp. specific primer of leptospiral major outer membrane protein LipL32 were highly exploited in previous studies[12,15-18].
Thus, this study aimed to screen and determine the infection of Leptospira spp. within R. rattus using a simplified molecular method of PCR-based diagnosis. The prevalence and genetic divergence of Leptospira spp. were investigated to address the following questions:(1) what is the prevalent patterns of Leptospira spp. infecting R. rattus,and (2) what is the degree of genetic divergence between Leptospira spp. based on 16S rRNA and LipL32 sequences. The outcomes of this study will contribute to the public health management and awareness of leptospirosis infection.
2. Material and methods
2.1. Ethics and permits approval
This study was endorsed by the Institutional Animal Care and Use Committee (IACUC), with the ethics reference no. ISB/10/06/2016/NHMI (R). The Department of Wildlife and National Parks authorised this study in collecting wild samples with permit reference number JPHL&TN(IP): 100-34/1.24 Jld. 7(6). This study was carried out without the involvement of any endangered species.
2.2. Study areas and samples collection
A total of 130 individuals of R. rattus (67 males, 63 females) from 13 cities in Peninsular Malaysia were enrolled in this study (Table 1, Supplementary Figure 1). The trappings of R. rattus were carried out with the help from municipalities of each city as part of their vector control program. Sampling was carried out at various sites(i.e. housing areas, fresh markets, and seaside) with emphasis on populated areas, as the study organisms are usually found nearby to human habitation. Trapping sessions were organised from December 2016 to February 2018 by using cage traps sized 28 cm × 15 cm × 12 cm.
The captured rats were identified based on their external size and features and were then confirmed, referring to the taxonomic keys[19-21]. Live specimens were euthanised in a sealed container connected to a carbon dioxide (CO2) tank. Dead specimens were then underwent dissection procedures after information such as external measurements, sex, and weight were recorded. Kidney and liver tissues were preserved in 95% (v/v) ethanol and kept in -80 ℃freezer before subsequent analysis.
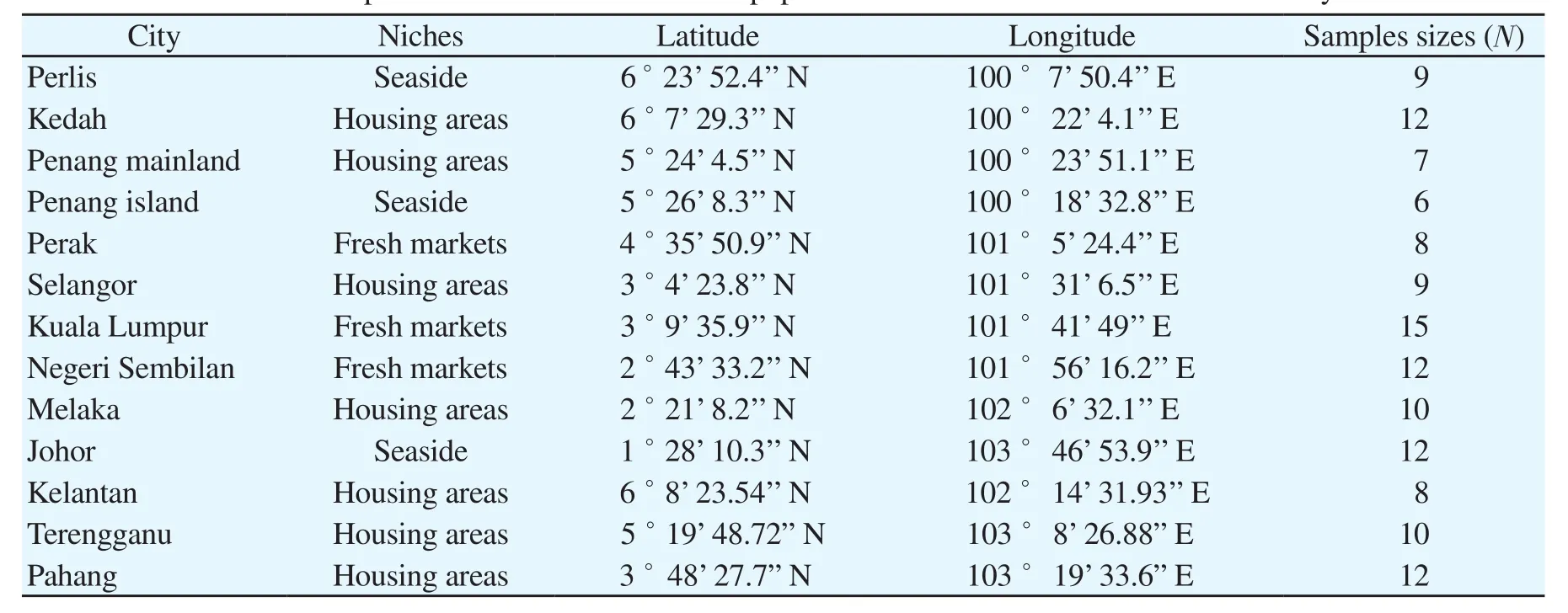
Table 1. Localities and samples sizes from which R. rattus populations were collected in Peninsular Malaysia.
2.3. DNA isolation and DNA amplification
Total genomic DNA from kidney and liver tissues were extracted following the protocols provided by the GF-1 Tissue DNA Extraction Kit (Vivantis Technologies Sdn. Bhd.). The purity of the extracted genomic DNA was determined by the A260/A280 nm ratio using 1:10 (DNA: buffer) dilution, with NanoDrop? 2000 Spectrophotometer (Thermo Scientific, USA).
The amplification of PCR was carried out using the PCR reagents included 12.5 μL of 2X Power Taq PCR MasterMix (BioTeke,Beijing), 0.5 μL of each 10.0 μM of forward and reverse primer,1.0 μL of genomic DNA template, and 10.5 μL of ultrapure water added up to the final volume of 25.0 μL, as suggested by the manufacturer. PCR was performed using two sets of established primers to amplify 412 and 240 base pair (bp) fragment correspond to 16S rRNA and LipL32 genes, respectively. The 16S rRNA primer is a Leptospira genus-specific primer, while the LipL32 is mainly for the pathogenic Leptospira spp. Primers Lep1 and Lep2 which related to 16S rRNA gene has forward sequence of 5'-GGAACTGAGACACGGTCCAT-3' and reverse sequence of 5'-GCCTCAGCGTCAGTTTTAGG-3'[22]. For LipL32 gene, the primer sequences are LipL32-2F (5'-TGGCTATCTCCGTTGCACTC-3')and LipL32-2R (5'-CCCATTTCAGCGATTACGGC-3')[23].Three reference strains of L. interrogans serovar Lai, L. interrogans serovar Copenhageni, and L. borgpetersenii serovar Javanica serve as positive controls, while bacteria of Escherichia coli acts as the negative control. Both control groups were used along in every PCR screening to eliminate most potential confounding results.
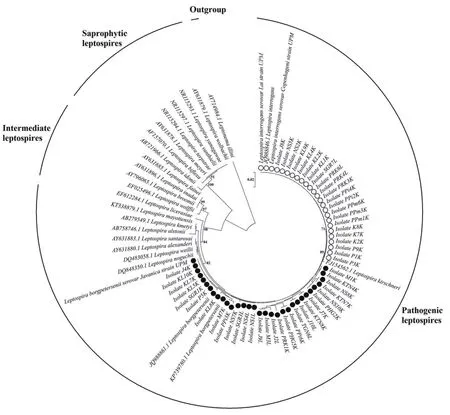
Figure 1. The neighbor-joining phylogenetic tree estimated using the Kimura-2-parameter algorithm and 1 000 bootstrap replications, based on 16S rRNA gene. Blank circles represent Leptospira interrogans while filled circles represent Leptospira borgpetersenii. The optimal tree with the sum of branch length= 0.3224 is shown and bootstrap values are indicated on the branches. The bar represents 0.02 nucleotide substitutions per alignment position.

Table 2. Average percentages of pairwise genetic distances among Leptospira spp. and Leptonema illini (Outgroup), based on the 16S rRNA gene.
DNA was amplified by using Mastercycler? Nexus (Eppendorf North America, Inc.). The profile for the PCR amplification were as follows: initial denaturation for four min at 94 ℃, continued with 35 cycles of denaturation for 30 s at 94 ℃, annealing for 30 s at 58 ℃, an extension for 30 s at 72 ℃, and a single cycle of a final extension stage for ten min at 72 ℃, before holding-up to 10 ℃ after the reaction completed. The PCR protocol designed for both 16S rRNA and LipL32 genes were same. The PCR products underwent electrophoresis using 2% (w/v) agarose gel in 1X TAE buffer. The pre-stained technique was used by adding SYBR? Safe DNA Gel Stain (Bioteke, Beijing) into the agarose gel. The gel electrophoresis was run at 80 V, 180 mA in 1X TAE running buffer for 45 min.The gels were then visualised under ultraviolet light (Alpha Imager Gel Documentation System, Siber Hegner, Germany). GF-1 PCR Clean-up Kits (Vivantis Technologies Sdn. Bhd.) were used to purify the DNA from PCR product, and the positive screening results were subsequently sent to Apical Scientific Sdn. Bhd. (Applied Biosystems 3730XL Genetic Analyzer) for nucleotide sequencing purposes.
2.4. Sequence and phylogenetic analysis
The 16S rRNA gene from 22 Leptospira spp. were used as reference sequences with the GenBank accession numbers as follows: FJ154562, JQ988846, DQ848350, JQ988861, KP739780,AY631880, DQ483058, AY631883, AB279549, AB758746,KT338879, EF612284, EF025496, AY796065, AY631896,AY631885, AB721966, AY631878, NR115294, AF157070,NR115297, NR115293, and AY631879. A single 16S rRNA sequence of Leptonema illini (AY714984) was used as an outgroup to root the phylogenetic tree. For LipL32 gene, only the sequences of L. interrogans (KY356961) and L. borgpetersenii (KT338940 and KT338941) were included. The nucleotide sequences of leptospires acquired from GenBank and those obtained from the kidney, liver,and positive control samples after the nucleotide sequencing process were aligned accordingly using the ClustalW multiple alignment algorithms of Molecular Evolutionary Genetics Analysis Version 7.0(MEGA7)[24]. The edited sequences were validated using sequence similarity searches (GenBank BLASTn) to ensure the targeted species locus sequences were obtained. The edited sequences were analysed using MEGA7 software. Phylogenetic tree was built in MEGA7 using distance-based neighbor-joining (NJ) tree. Haplotype sequences were evaluated from the DNA dataset using DNA Sequence Polymorphism Version 5.10.1 (DNASP v5)[25]. Network 5.0.0.1 was used to generate a minimum-spanning network (MSN)to illustrate the haplotype relationships[26].
3. Results
Both leptospiral 16S rRNA and LipL32 sequences were successfully amplified from the kidney and liver tissues of R. rattus, with expected amplicon size of 412 and 240 bp, respectively. Out of 130 individuals of R. rattus studied, a total of 51 (39.23%) individuals were positive for leptospirosis detected by using PCR method. The sequences amplified using both primers denoted that all the positive DNA were Leptospira spp. and belongs to the pathogenic leptospires (98%-100% maximum identity). Two pathogenic Leptospira spp. namely, L.interrogans and L. borgpetersenii were discovered circulating among R. rattus. The most dominant species is L. borgpetersenii carried by 29 (56.86%) individuals while another 22 (43.14%) individuals were colonised by L. interrogans. No intermediate and saprophytic leptospires species were identified in this study. The nucleotide sequences of all the positive controls and samples were deposited into GenBank database with accession numbers of MH997573-MH997626 for 16S rRNA gene and GenBank accession numbers of MK026014-MK026067 for LipL32 gene.
All of the captured R. rattus comprised 69 subadult and 61 adult rats. The leptospires infection, according to age group was quite similar with 39.10% and 39.30% occurrence in subadults and adults'rats, respectively. No juvenile rats were trapped in this study. Males R. rattus are the most prevalent to leptospires infection compared to females. Out of 67 males R. rattus, 28 (41.79%) individuals are the vectors for leptospirosis, in which 12 and 16 of them were infected with L. interrogans and L. borgpetersenii, respectively. From 63 females R. rattus, a total of 23 (36.50%) individuals were infected by leptospires, in which 10 and 13 individuals harboured L. interrogans and L. borgpetersenii, respectively.
Our sampling areas, mainly focus on three preferable niches of fresh markets, seaside, and housing areas where populations of R.rattus are abundant. Our results demonstrated that rats caught from fresh markets are the most prevalent with leptospires infection. Out of 35 rats caught in fresh markets, 19 (54.28%) of them are carriers of leptospires, in which 9 and 10 of them carrying L. interrogans and L. borgpetersenii, respectively. Meanwhile, of total 27 rats caught in seaside niche, 14 (51.85%) of them are infected with leptospirosis,wherein 6 of them were infected by L. interrogans and another 8 individuals corresponded to L. borgpetersenii. However, with the highest number of 68 rats trapped in the housing areas, only 18(26.47%) individuals harbouring leptospires. 7 and 11 of them carrying L. interrogans and L. borgpetersenii, respectively.
Furthermore, our study portrays the occurrence of leptospiral DNA in both kidney and liver tissues of R. rattus. Overall, from each kidney and liver tissues of the 130 rats utilised in the screening of infection, 11 (8.46%) liver tissues were colonised by Leptospira spp.,in which 3 individuals were infected with L. interrogans and another 8 individuals are in agreements with L. borgpetersenii. A total of 40(30.76%) individuals of R. rattus were proved leptospirosis positive after the kidney tissues were screened. 19 L. interrogans and 21 L.borgpetersenii species colonised the kidney of the rats. Surprisingly,none of the individuals got infected in both kidney and liver tissues simultaneously. The leptospiral DNA presented in either one of the two screened organs of an individual at each time point.

Figure 2. The neighbor-joining phylogenetic tree estimated using the Kimura-2-parameter algorithm and 1 000 bootstrap replications, based on LipL32 gene. Blank circles represent Leptospira interrogans while filled circles represent Leptospira borgpetersenii. The optimal tree with the sum of branch length= 0.0652 is shown and bootstrap values are indicated on the branches.The bar represents 0.005 nucleotide substitutions per alignment position.
For species identification using phylogenetic analysis, the Kimura-2-Parameter model was used in NJ tree reconstructions tested with a bootstrap value of 1 000. From 16S rRNA gene sequences, three well-defined clades of Leptospira spp. were successfully resolved(Figure 1), referred to the clades of pathogenic, intermediate, and saprophytic species. The leptospires DNA obtained from kidney and liver tissues of R. rattus were clustered into the pathogenic clades of L. interrogans and L. borgpetersenii. The positive control samples were also segregated accordingly. Essentially, the tree revealed monophyly of the genus Leptospira with respect to the outgroup, Leptonema illini. Phylogenetic comparisons were also initiated between the two Leptospira spp. using LipL32 gene. Figure 2 indicates that the similar tree topology was derived from all the sequences, in which two separated clades of L. interrogans and L. borgpetersenii were formed.The average percentages of pairwise genetic distances among Leptospira spp. and outgroup based on 16S rRNA gene calculated using Kimura-2-Parameter algorithm model is tabulated in Table 2.From the analysis, pathogenic leptospires of L. interrogans and L.borgpetersenii observed in this study demonstrated an interspecific genetic distance of 0.60% between them. Interestingly, the genetic distances between pathogenic, intermediate, and saprophytic groups of leptospires revealed the most prominent results. The relationships of the pathogenic group with intermediate and saprophytic groups are in the range of 1.70%-3.70% and 8.80%-10.20%, respectively.Species of genus Leptospira exhibited a highest value of a pairwise distance with Leptonema illini between 15.20%-18.00%, which were highly expected as they were originated from two different genera. From the calculation of pairwise distances, we may conclude that the genetic distances of 16S rRNA sequences can explain the relationship between the species and genus level. Additionally, the pairwise genetic distance between L. interrogans and L. borgpetersenii corresponded to LipL32 gene shows greater nucleotide divergence with the value of 5.80%. The result signified that 16S rRNA gene had a lower rate of base substitution and was more conserved as compared to LipL32 gene.
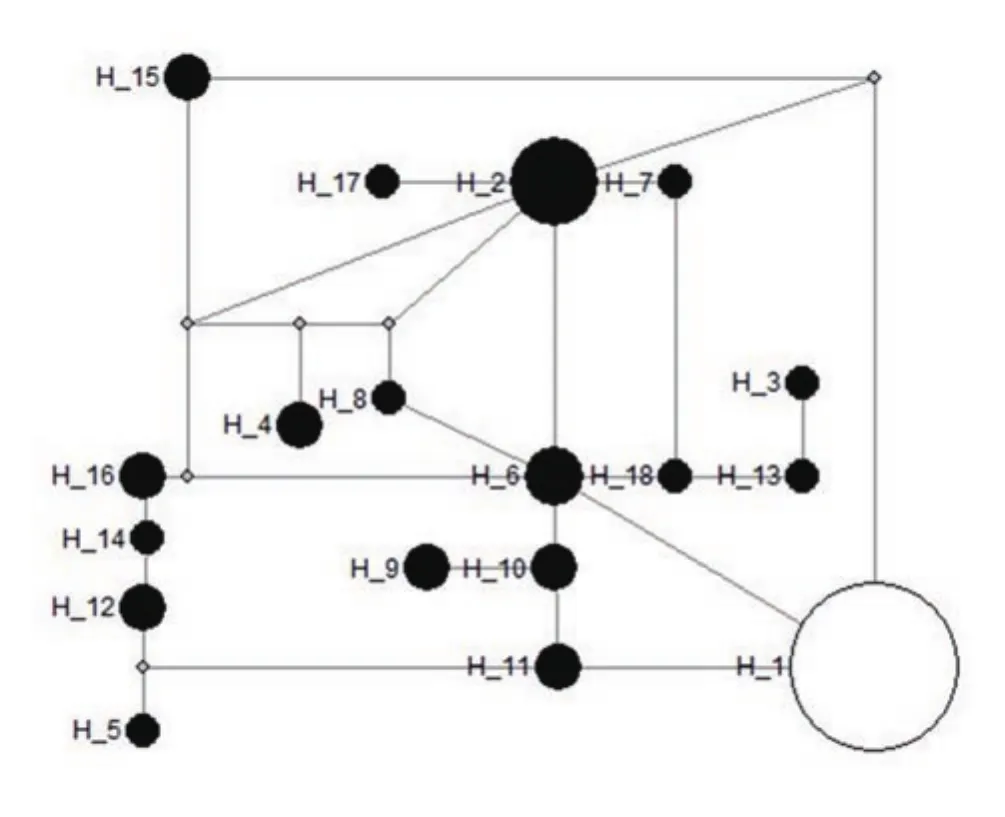
Figure 3. Minimum-spanning network of Leptospira interrogans and Leptospira borgpetersenii haplogroup defining 18 haplotypes, based on 16S rRNA gene. Haplotype 1 - Leptospira interrogans; Haplotype 2-18 -Leptospira borgpetersenii. The circle size of Leptospira spp. haplotypes is proportional to the frequency of the haplotype.
Single-nucleotide polymorphisms analysis conducted on L.interrogans and L. borgpetersenii sequences based on the 16S rRNA and LipL32 genes revealed 12 and 15 segregating sites, respectively.Throughout the analysed sequences of the two species, 18 haplotypes were defined for 16S rRNA gene and 5 haplotypes were derived from LipL32 gene. Minimum-spanning network was generated with the haplotype data obtained to illustrate the relationships of the two Leptospira spp. For 16S rRNA gene, L. interrogans solely colonised in a single haplotype (H_1) and the remaining 17 haplotypes were conquered by L. borgpetersenii (H_2-H_18) (Figure 3). Among 5 haplotypes detected using LipL32 gene, two haplotypes were observed for L. interrogans (H_1-H_2), as well as 3 from L.borgpetersenii (H_3-H_5) (Figure 4). The network analysis of both genes revealed that the haplotypes are unique to each species, as all of them lacked a common haplotype.
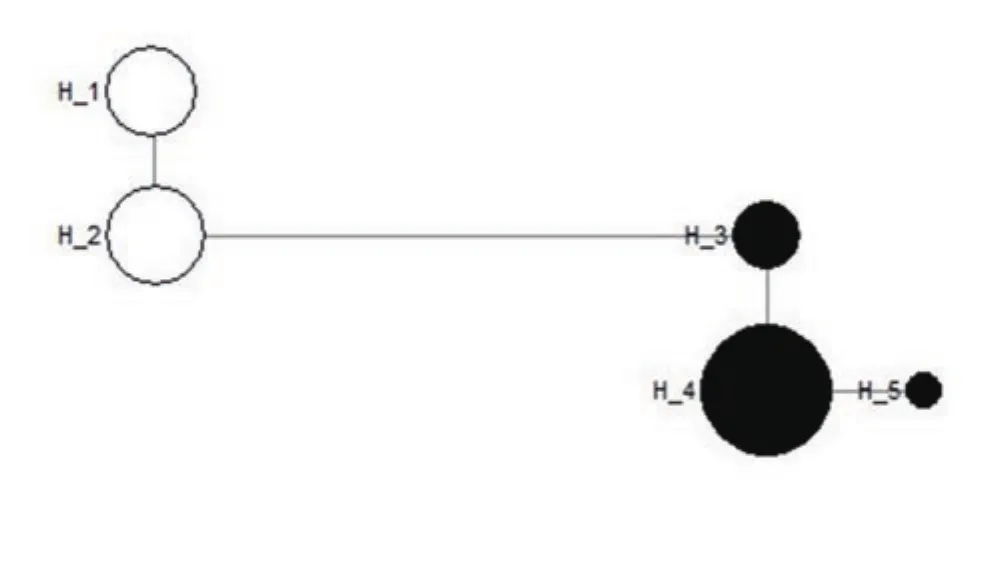
Figure 4. Minimum-spanning network of Leptospira interrogans and Leptospira borgpetersenii haplogroup defining five haplotypes, based on LipL32 gene. Haplotype 1-2 - Leptospira interrogans; Haplotype 3-5 - Leptospira borgpetersenii. The circle size of Leptospira spp. haplotypes is proportional to the frequency of the haplotype.
4. Discussion
Leptospires are globally distributed, however tropical and subtropical regions as Malaysia attracts the bacteria for longest survival[1]. Due to the diversity of the bacteria living in different environments, human and animals are exposed to a continuous risk of infection. From this study, the discovery of pathogenic Leptospira spp. in rats was predictable as previous study first documented the isolation of this spirochete from rats[27]. The findings of L.interrogans and L. borgpetersenii infecting rats is in agreement with earlier studies[12]. The two species pose a serious health risk as both of them are commonly known as the main causative agent of leptospirosis globally, which can lead to fatality[28]. The identification of disease reservoirs is paramount in assisting in the planning of effective prevention and control measures, limiting the proliferation rate of the spirochetes in the environment, as well as minimising the exposure risk of human comes into contact with the infected animals.
Molecular approach, in particularly PCR, is regards as a potent technique in detection of leptospirosis infection in rats as demonstrated in this study. The incorporation of both 16S rRNA and LipL32 protein genes in the screening not only detect the presence of the bacteria, but also differentiate between pathogenic and nonpathogenic Leptospira spp. In addition, by knowing the nature of leptospiral colonisation in an infected host, selective screening on both liver and kidney tissues reveals the disease's development stage,be it early or late infection, respectively. So, PCR holds as a rapid tool for diagnosis.
Out of 51 rats recognised as leptospiral positive, the prevalence of leptospires in subadult (39.10%) and adult (39.30%) rats do not differ significantly. The aggressiveness of both age groups in fighting for resources may have led to the transmission and circulation of Leptospira spp. among them. However, this study discovered that the infection is more prevalent in male rats compared to the females. This finding is in accordance with previous study as higher prevalence of Leptospira spp. was discovered in male rats[29]. This may be due to the different behaviour and lifestyle of both sexes in which females are confined within a smaller area in demands to pregnancy and lactation for the young, while males have higher frequency in encountering with others to contend for breeding territories and mates.
In furtherance, our sampling niches of fresh markets, housing areas,and seaside disclosed the pervasive distribution of rats within these areas. The rapid urbanisation and the emerging of a run-down area with insufficient facilities and poor sanitation bring the dispersion of rats closer to human and enhance the possibility of dissemination of rodent-borne diseases. Our results indicated that the rats caught in fresh markets were highly exposed to the leptospiral infection.The condition of the fresh markets with abundant resources for the rats to scavenge, the improper maintenance of the drainage system congested with rubbish, and the wet containers left exposed create a risk of feasible contamination with rat's urine. The moist surroundings promote the survival of pathogenic leptospires outside the hosts, aiding as a catalyst of leptospiral infection in both humans and animals.
R. rattus are mostly asymptomatic during the leptospiral infection and continue to shed the pathogen into the environment through urination. Our results demonstrated a higher occurrence of the pathogen in the kidney tissues compared to the liver tissues. An assumption can be made herein, where kidney is the most favourable place which promotes continuous colonisation of Leptospira spp.Previous study suggested that the proximal tubule of the kidney represents a secure place for Leptospira spp. to stay as they are shielded from the activity of the immune system[30]. The incident of the leptospires in the liver tissues are most likely to happen during the early stage of bacterial infection as the bacteria spread comprehensively to nearly all tissues throughout the body[31].However, with the production of antibody within days, the pathogens are rapidly eliminated from most tissues with the help of the immune defence, except the kidney[31,32].
The phylogenetic reconstructions of 16S rRNA gene revealed that the genetic make-up of all the three groups of Leptospira spp. (pathogenic, intermediate, and saprophytic) are unique and discriminative among them. The results of genetic distance also support the separation of the groups. In addition, the genetic constitution between both pathogenic species of L. interrogans and L. borgpetersenii disclosed a huge species variation. The comparisons of genetic distance between L. interrogans and L. borgpetersenii remarked LipL32 gene as the best tool for the recognition of pathogenic Leptospira spp. due to greater evolutionary divergence compared to 16S rRNA gene.
The results of genetic divergence of the two closely related species of L. interrogans and L. borgpetersenii indicated that several genetic variations existed within and between species, which further disclosed significant levels of genetic structure. It is surprising that our result of pathogenic Leptospira spp. identified from rats disclosed a strong relationship with Leptospira spp. spotted from another host, such as Sus scrofa[33]. This indicates that the Leptospira spp. inhabiting animals, humans, and environment were genetically identical and have a broad host range. To date, there is no proof that certain genetics composition only response to a particular host and reservoir[34].
Notably, the amplification of 16S rRNA gene highlighted the individuals of L. interrogans shows no variation among them,while individuals of L. borgpetersenii shows huge variation. This indicates that L. borgpetersenii is highly mutated and easily evolved,contributing to the sequence variations as compared to L. interrogans.However, through the amplification of LipL32 gene, the variation among individuals of L. borgpetersenii become less prominent and comparable with L. interrogans. The conserved region of LipL32 gene demonstrated a slower mutation rate within L. borgpetersenii.
Overall, this study presents an insight on essential surveillance information on the prevalence of Leptospira spp. from rats in Peninsular Malaysia. Two pathogenic Leptospira spp., namely L.interrogans and L. borgpetersenii were discovered from R. rattus with a large dissemination, emphasising a critical public health concern. To a greater extent, this study suggests that PCR method is a reliable tool in molecular diagnostic which succeeds in discovery of leptospiral DNA from rats. The diagnosis of leptospirosis can be facilitated using 16S rRNA and LipL32 genes as molecular markers of infection. Rats should be marked as the most important host and reservoir of Leptospira spp. The risk of leptospirosis infection can be diminished with numerous preventive measures such as rodent control program. The findings from this study could provide hints for subsequent molecular, clinical, and epidemiological research to execute advance diagnostic strategies.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Authors' Contributions
NHMI, SB, KS, SU, CCC, and HO contributed equally to the concepts, design, definition of intellectual content, literature search,manuscript editing, and manuscript review. NHMI performed the data acquisition, data analysis, and statistical analysis. Both NHMI and CCC involved in the experimental studies and manuscript preparation. All authors have read and approved the final manuscript.
 Asian Pacific Journal of Tropical Medicine2019年10期
Asian Pacific Journal of Tropical Medicine2019年10期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Non-related contact lens coinfection with Acanthamoeba and Fusarium
- Comparative analysis of current diagnostic PCR assays in detecting pathogenic Leptospira isolates from environmental samples
- Detection of Trypanosoma spp. in Bandicota indica from the Thai-Myanmar border area, Mae Sot District Tak Province, Thailand
- Molecular characterization and subtyping of Blastocystis in urticarial patients in Turkey
- Manifestations and outcomes of leptospirosis during local outbreaks in high endemic districts of Sri Lanka: A retrospective multi-center study
- Should pyridoxine be given to breastfed infants whose mothers are on isoniazid?
