Molecular aspects of eye development and regeneration in the Australian redclaw cray fish,Cherax quadricarinatus
Tomer Ventur ,Michel J.Stewrt,Jennifer C.Chndler,Bronwyn Rotgns,Abigil Elizur,1,Alex W.Hewitt
a GeneCology Research Centre,Faculty of Science,Health,Education and Engineering,University of the Sunshine Coast,Sunshine Coast,Queensland 4556,Australia
b Lions Eye Institute,University of Western Australia,Perth,Western Australia 6330,Australia
c School of Medicine,Menzies Institute for Medical Research,University of Tasmania,Hobart,Tasmania 7005,Australia
Keywords:Ocular development Regeneration Compound eye Crustacean Cherax quadricarinatus Transcriptome
A B S T R A C T The compound eye evolved over 500 million years ago and enables mosaic vision in most arthropod species.The molecular regulation of the development of the compound eye has been primarily studied in the fruit fly Drosophila melanogaster.However,due to the nature of holometabolous insects halting growth after their terminal metamorphosis into the adult form,they lack the capacity to regenerate.Crustaceans,unlike holometabolous insects,continue to grow during adulthood,achieved through regular shedding of their exoskeleton,in a cyclic process known as molting.This therefore offers crustaceans as a highly suitable model to study ocular regeneration in the adult arthropod eye.We have assessed the regenerative capacity of the retinal section of the Cherax quadricarinatus(red-claw cray fish)eye,following ablation and successive post-metamorphic molts.This work then provides a transcriptomic description of the outer,pigmented retinal tissue(the ommatidia and lamina ganglionaris)and the basal,non-pigmented neuroendocrine ocular tissue(the X-organ Sinus Gland complex,hemiellipsoid body and optic nerve).Using comparative analysis,we identified all the transcripts in the C.quadricarinatus ocular transcriptome that are known to function in compound eye development in D.melanogaster.Differentially and uniquely transcribed genes of the retina are described,suggesting proposed mechanisms that may regulate ocular regeneration in decapod Crustacea.This research exemplifies the application C.quadricarinatus holds as an optimal model to study the regulation of ocular regeneration.Further in-depth transcriptomic analyses are now required,sampled throughout the regeneration process to better define the regulatory mechanism.
1.Introduction
The compound eye evolved an estimated 520 million years ago,generating a complex tissue that enables mosaic vision in most arthropod species,which account for most animal species on earth(Clarkson,Levi-Setti,&Horvath,2006).The compound eye consists of several similar anatomical units known as ommatidia,which are encased on their exposed surface by a faceted,transparent,and multi-layered cornea(Meyer-Rochow,2001).Each ommatidium is composed of a cluster of photoreceptor and accessory cells,which together form the structural unit of the compound eye and the reflective layer of the retina(Cronin&Marshall,2001;Meyer-Rochow,2001).Aside from some select insect groups,where an intermediate form is found,the compound eye can be divided into two functional-structural arrangements known as the apposition and superposition eye(Nilsson,1983).While appositional eyes enable better resolution,the superposition eye has evolved in nocturnal and burrowing species as an adaptation to low light intensity(Meyer-Rochow,2015).In decapod crustacean species which often comprise planktonic larval stages and benthic,nocturnal post-metamorphic stages,the eye changes from appositional to superpositional as an adaptive response to the changes in environment and life style;an example being the spiny lobster,Jasusedwardsii(Mishra,Jeffs,&Meyer-Rochow,2006).To date,much of the research into the development of the compound eye has focused on the apposition eye(Friedrich,2003),employing the fruit fly Drosophila melanogaster as the model organism,with investigations devoted to studying the underlying molecular pathways that orchestrate its initial development(Silver&Rebay,2005).
The cell fate of the primordial eye in D.melanogaster is set by the time an individual reaches the second instar larva,through a biochemical cascade which progressively defines the entire head region.Competing transcription factors that are produced at different levels by different cells in the head region,define the boundaries of the eye from the antenna and head cuticle,leading to differential expression in the region that will become the adult eye(Treisman,2013).Autoregulatory loops in the cells committed to become the retina progressively drive photoreceptors differentiation.The genes responsible for ocular development in D.melanogaster include master control genes which are considered to function as a “transcriptional unit”,with the loss of any one resulting in malformation(Kumar,2001).The genes include eyeless(ey),twin of eyeless(toy),sine oculis(so),eyes absent(eya),as well as dachshund(dac)and optix which were shown to be sufficient to induce ectopic eye(Kumar,2001).In addition,there is an array of secondary genes that integrate into the complex regulatory network that coordinates ocular differentiation,one example being hedgehog(hh)which sets in motion a cascade that leads to step-wise differentiation of photoreceptors through the activating Spitz,a ligand for the epidermal growth factor receptor(EGFR)(Treisman,2013).Although these genes are not ocular-specific and tend to be involved in multiple cross-tissue developmental processes,the role of the key regulators ey and toy(known as Pax6 in vertebrates)in ocular development is conserved across Phyla,being functionally described in multiple vertebrate classes(Kumar,2001),as well as the Lophotrochozoa(Tessmar-Raible&Arendt,2003).
Interestingly,in Crustacea(specifically the giant freshwater prawn,Macrobrachium rosenbergii),EGFR gene silencing inhibited eye development as well as weight increment following consecutive molts during post-metamorphic growth(Sharabi,Ventura,Manor,A flalo,&Sagi,2013).This is perhaps due to the systemic effect of EGFR silencing,causing a general inhibitory effect to tissues which rely on EGFR for adequate development.In contrast to holometabolous insects(which cease growing once metamorphosed into the final adult form),crustaceans continue to grow throughout their adult life,requiring regular molts of their exoskeleton.These growth patterns offer Crustacea as an optimal model to study the regulation of post-metamorphic growth in Arthropoda.Perhaps more interestingly,these successive molts mean crustaceans are capable of regenerating lost or damaged limbs through the molt cycle(Chang&Mykles,2011;Hopkins,2001;Mykles,2001),including the eye.Therefore,crustacean species not only present a unique model for studying the development of the compound eye over a transitional life-cycle,they also offer the unique opportunity to investigate ocular regeneration.
To date,studies describing regeneration in Crustacea have focussed on limb regeneration,which occurs in response to limb loss or damage,coordinated through the molt cycle(Chang&Mykles,2011;Hopkins,2001;Mykles,2001;Skinner,1985;Skinner&Graham,1970).The endocrine pathways underlying limb regeneration have been well characterised,involving fibroblast growth factors(FGF)and ecdysteroids,which function through a nuclear receptor heterodimer,comprised of the Ecdysone Receptor(EcR)and Retinoid Receptor(RXR)(Das&Durica,2013;Hopkins,2001).Together,this work highlights that the endocrine system that coordinates molting in Crustacea,also regulates limb regeneration,as the two processes are tightly integrated.For example,silencing of the EcR/RXR heterodimer in the blastema of the regenerating limb has been shown to cease celldivision in the limb bud,halting the regenerative process,whilst also resulting in the systemic effect of failure to molt(Das&Durica,2013).Interestingly,the circulating ecdysteroids that coordinate these processes are primarily regulated by a neurohormone released from the eyestalk(specifically from the neuroendocrine complex known as the X-organ Sinus Gland complex(XO-SG)(Chang&Mykles,2011;Mykles,2001;Skinner,1985)).The neurohormone known as Molt Inhibiting Hormone(MIH)inhibits the production of ecdysone by the Y-organs,meaning eyestalk ablation allows for an ecdysteroid peak and consequential precocious molt;an effect also stimulated by limb loss(Mykles,2001).Although not in Decapoda,the genetic basis of this cellular regeneration has been investigated in the amphipod crustacean,Parhyale hawaiensis. This work has highlighted the role of lineage-committed progenitor cells,suggesting a conserved mode of muscle regeneration with the vertebrates(Konstantinides&Averof,2014).However,while the regulation of regeneration itself has been investigated,the role of cellular reprogramming as part of the regeneration process has received little attention.In the context of ocular regeneration,data is limited to a handful of observational studies from over a century ago(Steele,1907)and more recently in a case described in the commercially important black tiger prawn Penaeus monodon,in the context of inducing gonad maturation(Desai and Achuthankutty 2000a).
Although there is a paucity of knowledge regarding the biochemical machinery that regulates regeneration in invertebrates,the regenerative capabilities of the eye have been explored in detail in the vertebrates(Goldman,2014).In particular the cellular reprograming that leads to the functional replacement of damaged retinal cells has been studied in several teleost fish(Lindsey&Powers,2007;Lindsey&Powers,2007;Sherpa et al.,2008).In these species,the regenerative Müller glial cells,are capable of genetic reprogramming to develop as one of any of the retinal cells,allowing for the specific migration,proliferation and differentiation of each cell type,ultimately enabling complete restored vision in the damaged eye(Lindsey&Powers,2007;Sherpa et al.,2008).
The firststep,cellular reprograming,was found to be initiated through the secretion of several factors,some of which appear to be specific to the regeneration process.In contrast,the secondary stages of cellular proliferation,migration and differentiation tend to be regulated through the ubiquitous pathways that are responsible for the regulation of primary eye development(Goldman,2014).The specific factors involved in cellular reprogramming in Chordata include a heparin-binding EGF-like growth factor(Hbegf)and a tumor necrosis factor-alpha(Tnf-α).Other transcription factors with a conserved function in retinal neurogenesis include the basic helix-loop-helix(bHLH)genes,aka Atonal(ato)in D.melanogaster,Math5 in Mus musculus and Xath5 in Xenopus laevis;although their exact function varies across species(Brown et al.,1998).
We have therefore harnessed the unique regenerative capacity of the crustacean eye,to expand our understanding of ocular regeneration.To do so,we have turned to the aquaculture species,Cherax quadricarinatus(red-claw cray fish).As an aquaculture species,C.quadricarinatus is readily abundant and has previously well-established research pipelines through which to synergistically tackle wider-reaching questions.This offers the species(as well as many other cultured Crustacean species)as a commercially and logistically viable research model.Herein,we report the regenerative capacity of the retinal section of the C.quadricarinatus eye,following eye-ablation through to full regeneration,through a series of successive post-metamorphic molts.In an attempt to elucidate the molecular mechanism coordinating this regeneration,we then report a transcriptomic analysis of the C.quadricarinatus ocular tissue,comparing the differentially and uniquely transcribed genes defining the outer(pigmented)retinal section(comprising the ommatidia and lamina ganglionaris)(Meyer-Rochow,2015)and the basal(non-pigmented)ocular tissue(comprising XO-SG,the hemiellipsoid body,optic nerve and 4 neuralcellclusters)(Skinner&Graham,1970).Finally,we considerthe mechanism through which ocular regeneration is regulated in this crustacean.
2.Materials and methods
2.1.Animals
C. quadricarinatus juveniles (weighing 0.86-2.59 g) were purchased locally and placed in separate fenestrated containers(16 cm×8 cm ×7.5 cm), floated in a 300 L aquarium with constant aeration and water filtration.Water temperature was maintained at 26±2°C.After a minimum 48 h of acclimation,the dioptric region,clear zone and retina were removed from the left eye while the right eye was removed entirely,including the neuronal ganglia and the XO-SG complex.Prior to dissections,animals were anesthetized in ice cold water for 10 min.Mature C.quadricarinatus individuals(23 males,weighing 49-104 g;27 females,weighing 49-120 g)were purchased and anesthetized similarly,from which the eyestalks were dissected and separated from the cuticle.
2.2.Histological analysis
Post the regeneration period,untreated and regenerated eyes were dissected as previously described.Eyes were rinsed in PBS and fixed in Bouin's solution(Sigma-Aldrich)for 16-24 h,then transferred to 70%Ethanol and stored at 4°C,until dehydrated and processed for sectioning and hematoxylin and eosin staining,as described in Ventura,A flalo,Weil,Kashkush,and Sagi(2011).
2.3.Sample preparation and sequencing
Male and female eyestalks were separately pooled,then placed in RNA-later(Ambion)for 24 h at room temperature,followed by storage at-20°C.From each pool,at least 20 eyes were taken for dissecting the retina(pigmented region)from the ganglionic region(non-pigmented region),generating a total of four samples(male and female;pigmented and non-pigmented regions)for sequencing.Dissection was performed based on visual observation of the pigmented region.Total RNA was isolated with the RNAzol RT Reagent(MRC),according to the manufacturer's instructions.Samples were sequenced by BGI(HongKong Co.Ltd)as per manufacturer's protocol(Illumina,San Diego,CA).Brie fly,poly(A)mRNA was isolated using oligo(dT)beads and the addition of fragmentation buffer for shearing mRNA into short fragments(200-700 nt).This was followed by cDNA synthesis using random hexamer-primers in order to prevent priming bias.The short cDNA fragments were further purified using QiaQuick PCR extraction kit and resolved with EB buffer for ligation with Illumina paired-end adapters.This was followed by size selection(~200 bp),PCR amplification and Illumina sequencing using an Illumina Genome Analyzer(HighSeq 2000,Illumina,San Diego,CA),performing 90 bp-paired end sequencing.The sequence reads were stored as FASTQ files,generating at least 4 GB of cleaned data(at least 45 million reads for each of the four samples).
2.4.Bioinformatics analyses
Cleaning of low quality reads,followed by assembly and mapping were conducted using the CLC Genomics Workbench(CLC Bio,version 7.0.3)under default parameters(with the exception of similarity fraction elevated to 0.9 in the mapping stage).From each library,69.3%-73.5%of the reads mapped to the complete de novo assembled C.quadricarinatus ocular transcriptome.BAM files were then uploaded onto Partek Genomics Suite(Partek GS)to quantify expression,defined as reads per kilobase of the transcript,per million reads of the total library(RPKMs).The RPKM values werefiltered to include only transcripts where RPKM≥1 in at least one sample.This subset of data was normalized to include a minimum of 0.05 instead of 0 RPKM and the normalized RPKMs were subjected to ANOVA,performed in Partek GS,to compare between the pigmented and non-pigmented samples.The threshold for statistical significance was set to P<0.005 and a fold-change of at least two was considered as significant.
To assess the level of conservancy with other arthropod eye development,we utilized the curated list of proteins annotated as functioning in D.melanogaster compound eye morphogenesis(Gene Onthology,GO identifier 0001745).To identify protein homologues in our C.quadricarinatus ocular transcriptome,we conducted a BLAST search of these eye development proteins against the predicted ORFs of our transcriptome(as predicted by ORF-Predictor http://proteomics.ysu.edu/tools/OrfPredictor.html),using CLC(v 7.5.1).Where homologous transcripts were identified,the amino acid sequences were aligned using the LaTeX TexShade package(Beitz,2000)and ORF and PFAM domain schematics were constructed using simple modular architecture research tool(SMART)(Schultz,Milpetz,Bork,&Ponting,1998).All reference sequences for phylogenetic analyses were retrieved from GenBank and trees constructed using MEGA(v 7.0)maximum likelihood.Con fidence levels for the groups defined in the topology were assessed by bootstrap and interior branch tests(with 1000 replicates).
3.Results and discussion
3.1.Eye regeneration assessment
Ten of the 22 individuals(45%;from which one eye was removed at the base,including both pigmented and non-pigmented tissue and the other eye,which was dissected to remove the pigmented retina only),survived for at least twelve weeks and completed three successive molts.Of these ten individuals,only those eyes where the retina alone was removed,were capable of partial regeneration in four individuals(Fig.1C,left eye and Fig.1D).None of the eyes that were removed at the base showed any evidence of regeneration over the three molts(Fig.1C,right eye).Interestingly,these observations differ slightly from that described in the penaeid prawn,P.monodon,where the authors describe the complete regeneration of fully ablated eyes,when both regions(pigmented and non-pigmented)were removed(Desai and Achuthankutty 2000b).However,as this observation was not a controlled experiment and rather a by-product of commercial eyestalk ablation,it may be that the entirety of the non-pigmented ocular tissue was not removed with complete accuracy.
Regeneration in C.quadricarinatus occurred as follows:by the first molt,a melanised scab developed around the removed retina,as has previously been documented in both ocular(Steele,1907)and limb(Das&Durica,2013;Hopkins,2001;Konstantinides&Averof,2014)regeneration in several crustacean species,suggesting a shared physiological regeneration response.By the second molt,the eye either failed to regenerate(n=4;40%),partially regenerated(n=4;40%;Figs.1 and 2),or regenerated in the form of an antenna(n=2;20%;Fig.2B).In those individuals that showed partial ocular regeneration at the second molt,by the third molt a seemingly completely regenerated eye was present.In these cases,although complete regeneration had occurred,comparative to untreated eyes(Fig.1A and B)the regenerated eyes were smaller(Fig.1C),with irregularities in the cornea surface(Fig.1D).These developmental abnormalities appear to be a common feature of ocular regeneration,as has been described in the zebra fish,where~50%of regenerated eyes showed defects and abnormalities and in some cases,lacked the ability to determine when regeneration was complete,resulting in over-proliferation(Sherpa et al.,2008).This suggests that even in those species that can achieve full ocular regeneration,the regenerative process does not achieve the optimal conditions of initial development.
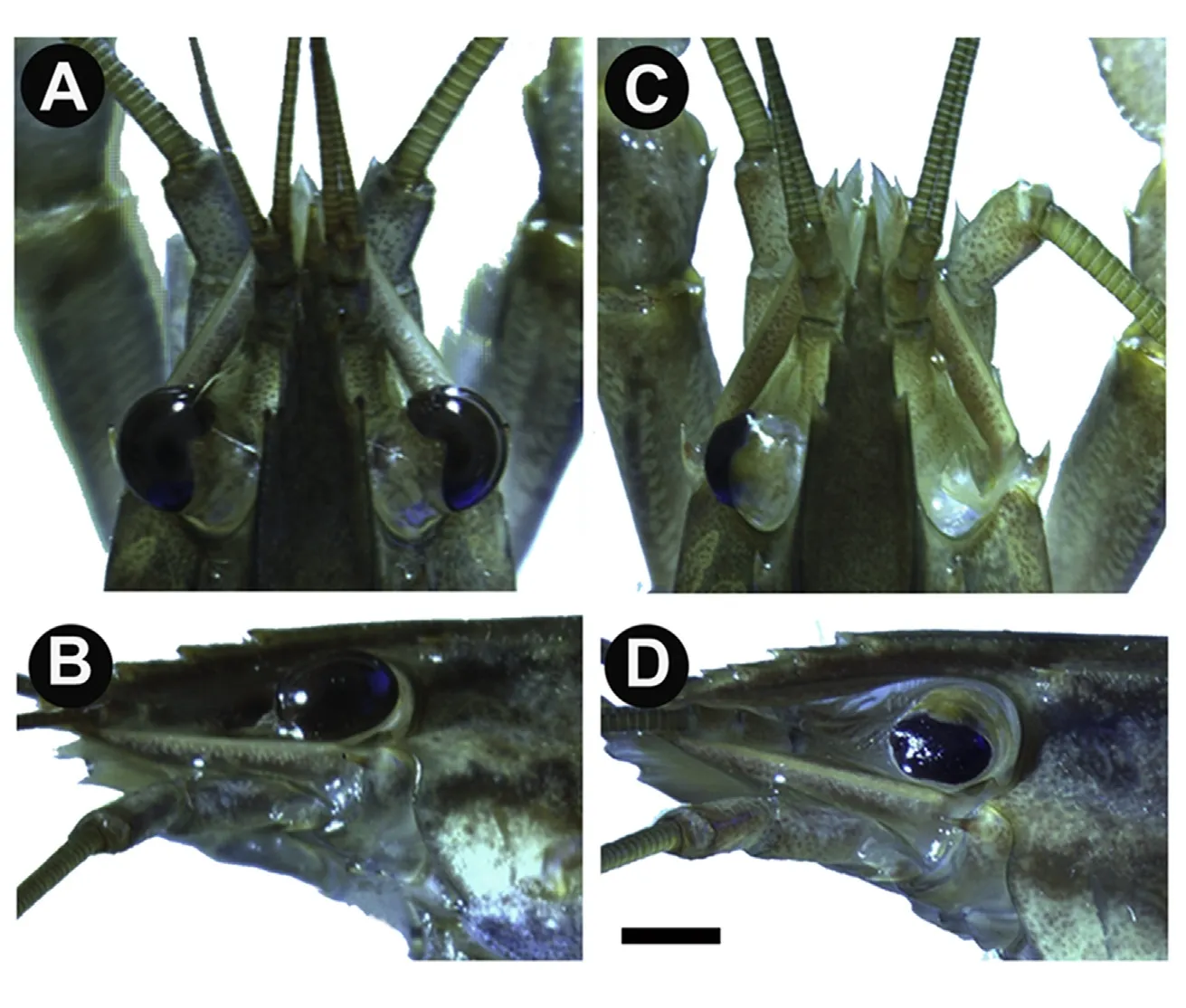
Fig.1.Complete eye regeneration in C.quadricarinatus.A)Dorsal view of an individual C.quadricarinatus with untreated eyes.B)Left side view of the same individual showing an intact eye with retina.C)Dorsal view of a C.quadricarinatus individual with full ablation of the right eye and developing retina in the left.D)Third-molt individual with regeneration of the treated left eye.Bar=5 mm.
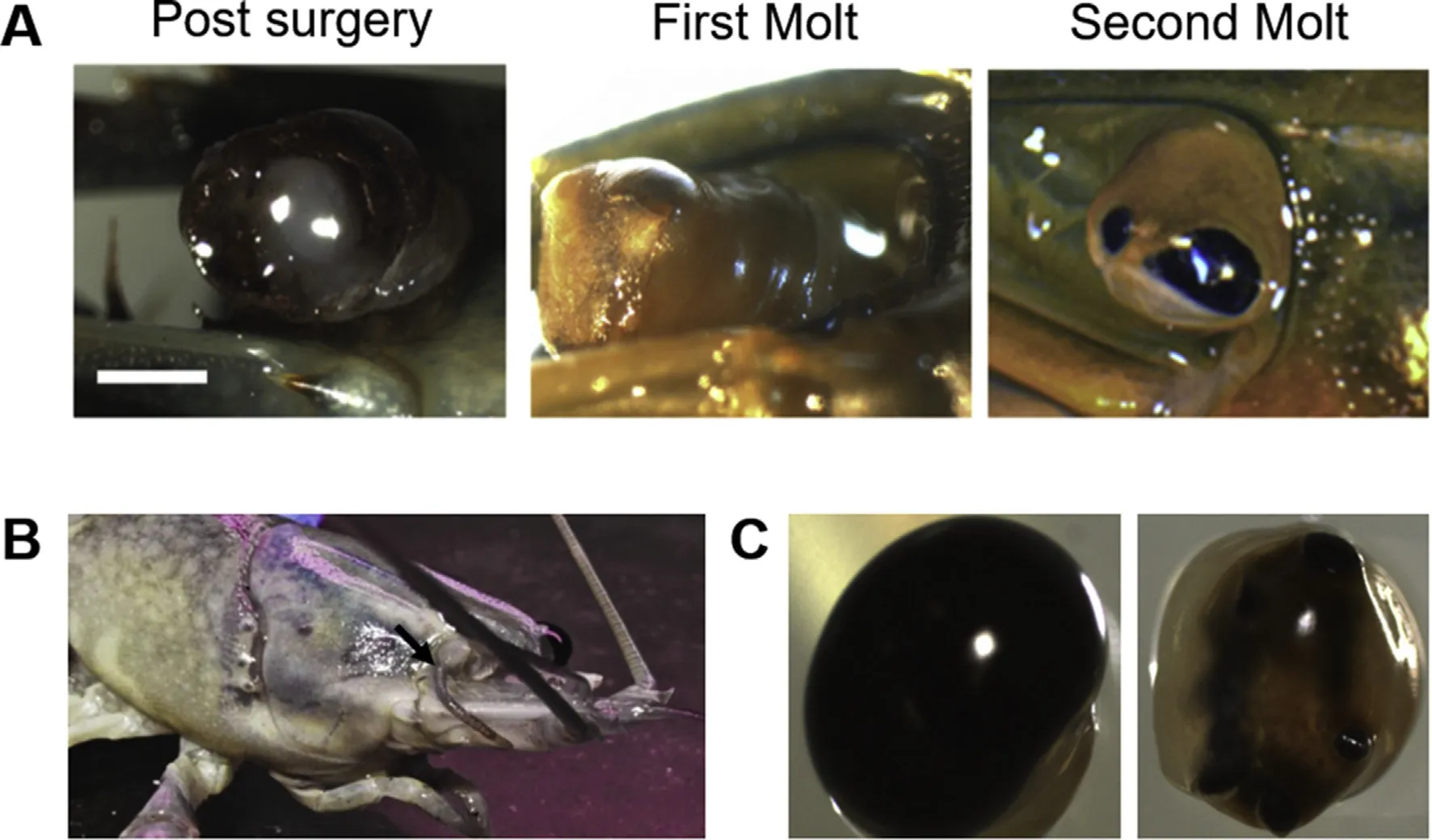
Fig.2.Varying levels of eye regeneration in C.quadricarinatus.A)The stages from post-surgery(left),showing removal of the retina(ommatidia and lamina ganglionaris),followed by formation of a melanised scab within one molt(middle)and partial regeneration of the retina within two molts(right).Bar=5 mm.B)Example of regeneration deformity where an antenna has regenerated in the retinal zone(highlighted by an arrow).C)An intact eye(left)and an incomplete regenerated eye(right),highlighting the irregularities of the regenerated eye.
One well-documented case of defective regeneration,is antennal regeneration in replacement of a damaged eye in D.melanogaster larvae.This was initially hypothesized to be due to the hyper-activation of the antennal-producing EGRF pathway,relative to the suppression of the ocular-inducing Notch pathway,stimulating a biochemical transition in development(Kumar,2001;Kumar&Moses,2001).It has been suggested that the specific effects of each pathway are,to an extent,shaped by the genetic background in which they function(Kumar,2001;Kumar&Moses,2001),although this model of Notch-EGFR eye-antenna specification has not been gaining support,considering the examples of antennal regeneration observed here(with complete development of all antennal substructures;Fig.2B),the “background”expression profile of an adult regenerating tissue,would probably differ quite dramatically from that of primary differentiating embryonic tissue and may well explain the emergence of developmental irregularities like that observed here,as well as that described in the zebra fish(Sherpa et al.,2008).A more likely mechanism for double-antenna discs generation in D.melanogaster showed that down-regulation of the Notch pathway results in a duplication of the antenna,rather than a switch in fate,as a consequence of a severe reduction in proliferation(Kenyon,Ranade,Curtiss,Mlodzik,&Pignoni,2003).With that,it is noteworthy that crustaceans show continual growth and in this study,we witnessed a fully differentiated eye which regenerated as an antenna,rather than the case in the holometabolous insect D.melanogaster where the disc which is in the process of differentiation has re-differentiated.More recently,a study by Wang and Sun(2012)has shown specific antagonistic pathways which define the eye and the antenna regions during the mid-second instar larval stage.A follow-up study examining the change in expression of these specific antagonistic factors in re-differentiating eyes to antennae in crustaceans could better explain the mechanism which enables a fully differentiated eye to regenerate as an antenna.
In the regenerated eyes,regeneration appeared to originate from the periphery of the retinal tissue.This is suggestive of two putative regenerative origins:either that these regionscontain remaining retinal cells as the source of regeneration,or the adjacent non-pigmented tissue contains the necessary progenitor cell clusters.The first case would suggest a regenerative mechanism in keeping with vertebrate studies,where the regeneration of all dioptric layers of the retina develop from the redifferentiation and proliferation of remaining retinal cells(Hitchcock,Lindsey Myhr,Easter,Mangione-Smith,&Jones,1992).The second case suggests a mechanism similar to limb regeneration studies described in the amphipod crustacean P.hawaiensis,where regeneration is achieved through lineage-committed progenitor cells,local to the amputated region(Konstantinides&Averof,2014).Considering the lack of regeneration with complete ocular ablation,it may be that the adjacent non-pigmented ocular region of the eye houses these locally sourced progenitor cells(or alternatively,cells which produce the molecular signal/s required for retinal cell reprogramming or retinal stem cell differentiation),which when removed,result in the lost regenerative capacity observed here(Fig.2C).We therefore sought to characterize the transcriptomic patterns that distinguish the pigmented and non-pigmented regions of the eye,in order to identify any key factors whose differential expression might implicate them in regulating the eye regeneration.
3.2.Transcriptome characteristics
Eye tissue from male(n=23)and female(n=27)C.quadricarinatus were separated into pigmented and non-pigmented regions and sequenced as such,generating over 180 million reads,which were de novo assembled to generate a total of 89,626 transcripts(mean length=706,N50=771).A total of 56,920 transcripts had an RPKM(reads per kilobase per million reads)of≥1 in at least one sample.
3.3.Conservation with Drosophila eye development
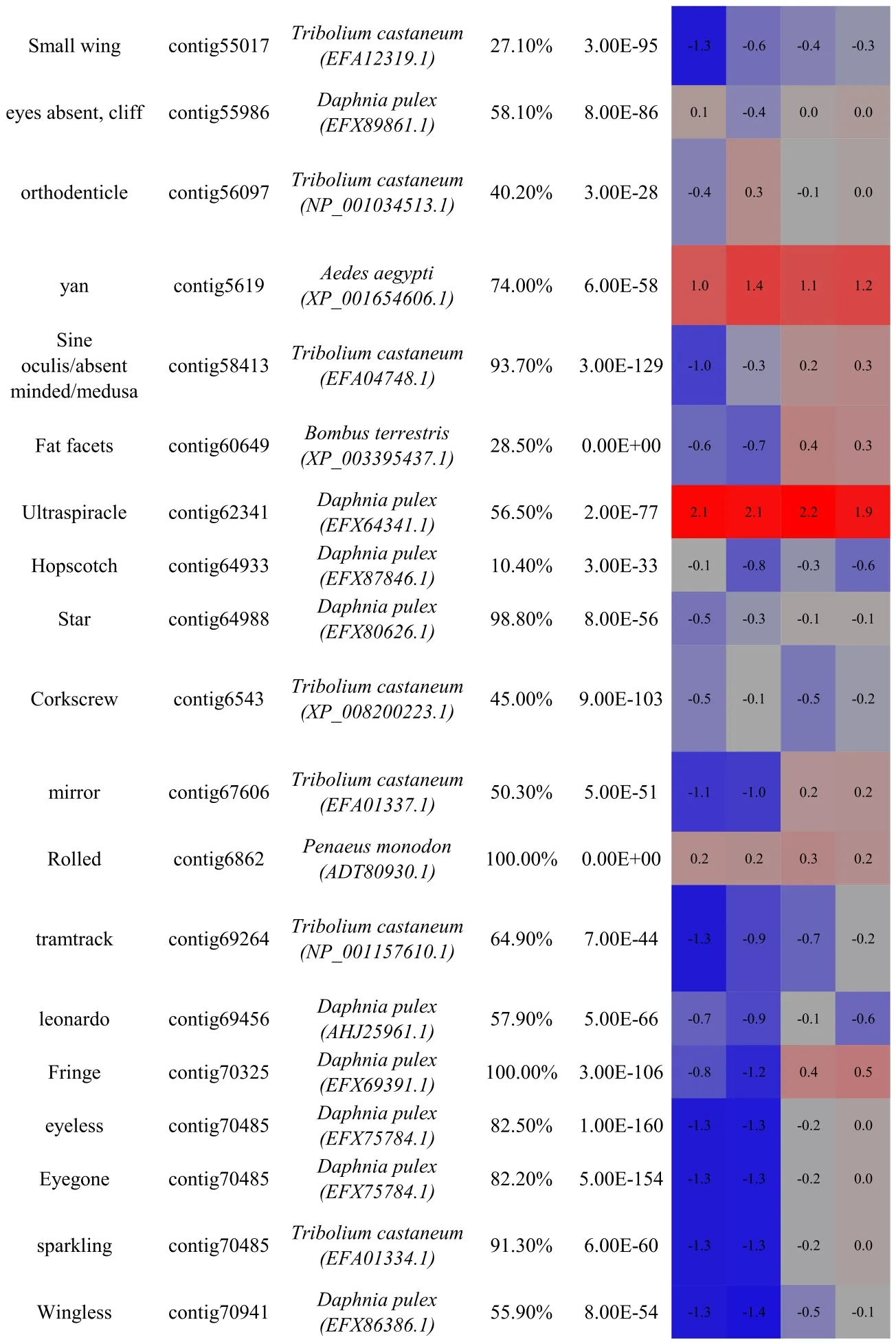
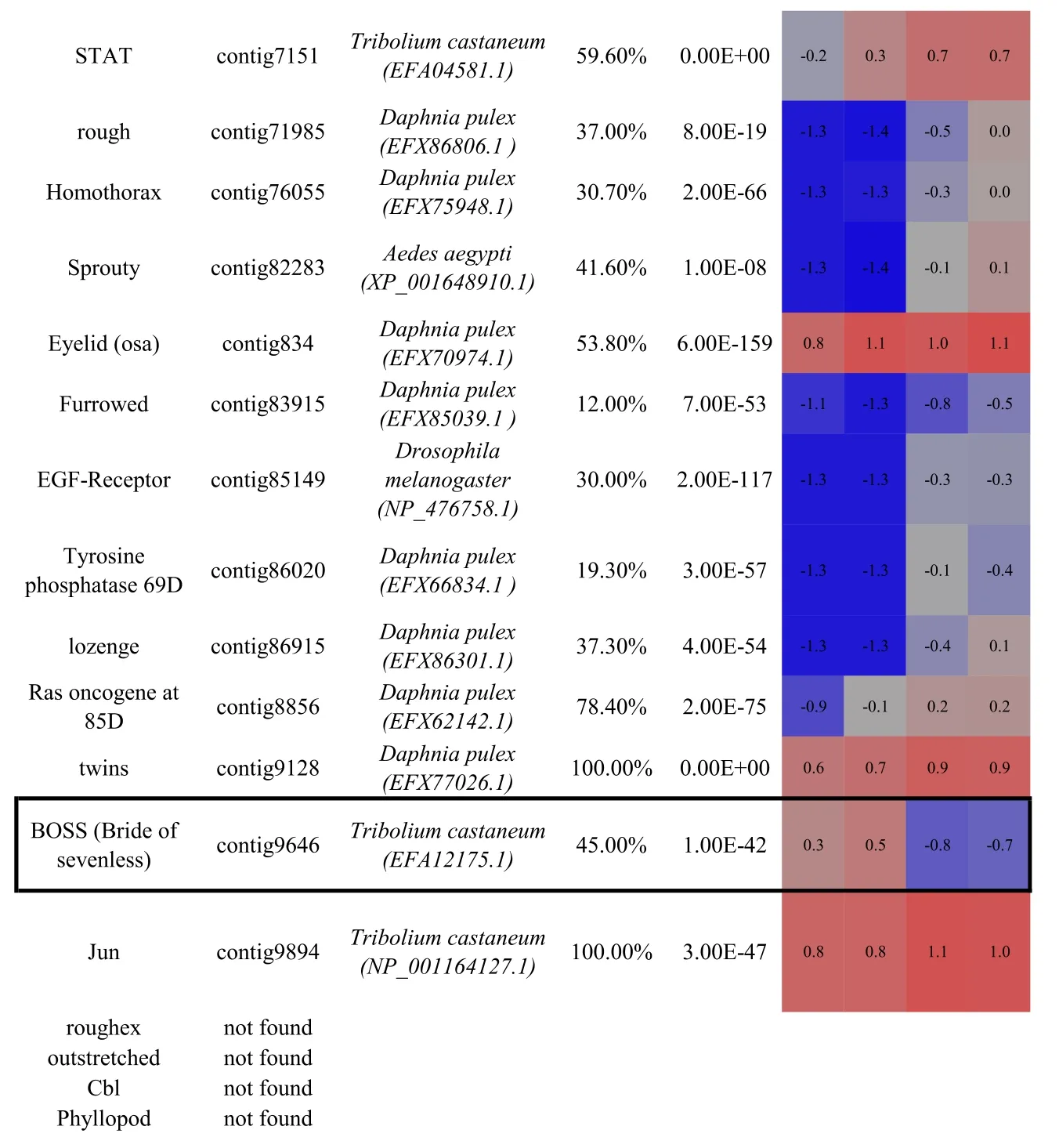
Of 469 proteins annotated to be involved in compound eye development in Drosophila,89%(418 proteins)were identified with an evalue≤10-5in the C.quadricarinatus ocular transcriptome(Table S1).The majority(50 out of 54)of proteins characterised as having eyedevelopment associated expression profiles in Drosophila(selected from the interactive fly,http://www.sdbonline.org/sites/ fly/aimorph/eye.htm),were also present in the C.quadricarinatus transcriptome;the four exceptions being roughex,outstretched,Cbl and Phyllopod.Of the 50 C.quadricaritatus homologues,most transcripts(when blasted at NCBI)shared highest similarity with proteins from the branchiopod crustacean,Daphnia pulex(29/50 factors)and the arthropod beetle,Tribolium castaneum(15/50 factors);Table 1.These annotations re flectthe availability of annotated genomes for D.pulex,T.castaneum and D.melanogaster and the phylogenetic distance each share with C.quadricarinatus.In summary,it is apparent that the vast majority of functional regulators characterised in eye morphogenesis in D.melanogaster,are present in the C.quadricarinatus ocular transcriptome.
Although the genetic basis of eye development is most extensively characterised in D.melanogaster,studies in other invertebrates have begun to extend this knowledge(Lapan&Reddien,2012;Tessmar-Raible& Arendt,2003).It appears that while some genes such as D.melanogaster's eyeless(ey)and twin of eyeless(toy),aka pax6 in vertebrates,are highly conserved in eye development across the nemertines,cephalopods,Dugesia,and Platynereis(although interestingly not in the planarian,Schmidtea mediterrane(Lapan&Reddien,2012))many of the other genes are not so conserved(Tessmar-Raible&Arendt,2003).This highlights the importance of species-specific characterisation.Planarians,as members of the Lophotrochozoa,the sister group to the Ecdysozoa(comprising all Arthropoda),are significant to understanding the molecular basis of eye evolution;furthermore,they are able to show full ocular regeneration capacity through adulthood(Lapan&Reddien,2012).RNA sequencing studies in the planarian,S.mediterrane,highlighted that while the(otherwise)highly conserved toy/pax6 gene family does not play a role in eye development,ocular development is instead regulated by a zinc finger transcription factor termed Ovo(Lapan&Reddien,2012).An ovo-like homologue is present in D.melanogaster(termed shavenbaby)but has only been studied in the context of epidermal morphogenesis,not eye development.Interestingly,the coordination of regeneration in S.mediterrane employs the same suite of genes(ovo,six-1/2 and eya)involved in initial embryonic development(Lapan&Reddien,2012).This suggests that regeneration occurs through re-establishing the genetic environment of embryogenesis.
3.4.Eye regeneration pathways
Other than that described in the planarian,the majority of studies regarding ocular regeneration have been conducted in the vertebrates.In vertebrates,eye regeneration is initiated and mediated by Müller glial cells.Although coordinated through different genes,regeneration to full visual capacity is achieved through the reactivation of the embryonic“developmental niche”(Sherpa et al.,2008),similar to that described in S.mediterrane(Lapan&Reddien,2012).The genes responsible in the vertebrates are hbegf and tnf-α9,which have a specific function in the cellular reprogramming and trans-differentiation of the Müller glia cells(Goldman,2014),rendering them pluripotentand thus recapitulating the developmental conditions of embryonic development(Sherpa et al.,2008).Although we were able to identify most components of the key Wnt and Notch regulatory pathways in the C.quadricarinatus ocular transcriptome,the master regulatory factors,tnfa and hbegfa,were not identified.Indeed,homologues of these genes could not be found in any Achordate when screened at NCBI,with the exception of a tumor necrosis factor(TNF)found in the shrimp species Litopenaeus vannamei.However,as this TFN is not a Tnf-α,it is unlikely to be functionally associated with retinal-cell reprogramming(Wang et al.,2012).This suggests that the same genetic coordination of cellular reprogramming as described in the vertebrates,is not conserved in Crustacea.
3.5.Pigmented and non-pigmented regions differential expression
Thus,in order to identify,as yet,uncharacterized factors with putative involvement in ocular regeneration in Crustacea,we conducted a transcriptomic comparison of the expression profiles of the pigmented(retinal)and non-pigmented(neuroendocrine)regions of the eye.Those transcripts with an RPKM≥1 in at least one of the four samples were subject to ANOVA.A total of 1325 transcripts had≥2 fold-change in expression between the two tissue types(P 0.005),of which 162(12.2%)were up-regulated in the pigmented(retinal)region(Fig.3A).Among the 50 key factors associated with compound eye development(Table 1),29 were up-regulated in the non-pigmented(neuroendocrine)tissue and only 3 were up-regulated in the pigmented(retinal)tissue(boxed in Table 1).
This expression pattern may re flect the diverse functional role of the non-pigmented region of the eye,which comprises musculature and connective tissues but more significantly,the major neuroendocrine XOSG complex(Skinner,1985).Furthermore,considering that this region must remain intact for retinal regeneration to occur,the non-retinal tissue may be that responsible for expressing the key genes involved in stimulating regeneration.This may be achieved as previously suggested,with this region housing the localised progenitor cells that serve as the source of regeneration(as described in P.hawaiensis(Konstantinides&Averof,2014)),as well as the XO-SG,responsible for the secretion of a suite of neuropeptides which govern a wide array of functions in crustaceans(Nguyen,Cummins,Elizur,&Ventura,2016),including the MIH and thus the more systemic regulation of molting and consequential ability to regenerate(Das&Durica,2013).
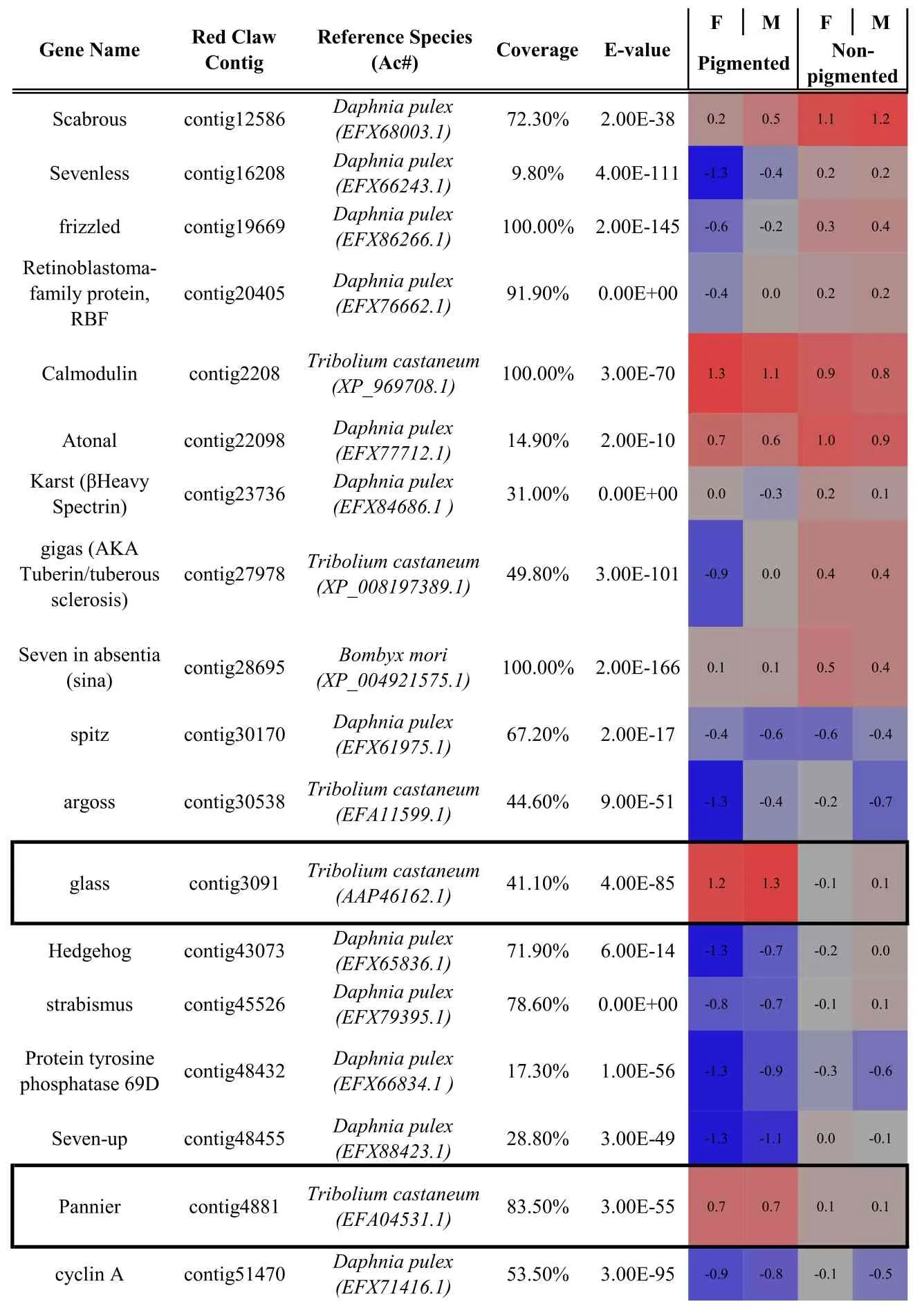
Table 1 Identification and differential expression of key genes known to be involved in compound eye morphogenesis(as characterised in model taxa)in the C.quadricarinatus eye transcriptome.Bold indicates the master control genes defined in Drosophila.Gene name,C.quadricarinatus eye transcriptome contig number,the species with highest similarity in sequence(and the protein accession number in parentheses),sequence coverage and E-value are shown.Coloured cells represent differential expression between the pigmented and non-pigmented regions of the eye(blue=down regulation;red=up-regulation;F=female;M=male).Up-regulated genes in the pigmented region(glass,pannier and BOSS)are boxed.Up-regulated genes in the non-pigmented region are underlined.For full expression details,refer to Table S2.
In contrast,if it is that regeneration originates from remaining retinal cells,still present after retinal amputation(in a mechanism similar to the vertebrate Müller glial cells),the factors responsible for the cellular reprogramming would be present and up-regulated in the pigmented region.Of the 162 transcripts identified as up-regulated in the pigmented region,only 28 had a predicted open reading frame(ORF)of 100 amino acids(aa)or more.This suggests either that most transcripts are truncated,or perhaps indicates the presence of long non-coding RNAs,known to be key players in cellular regulation(Mercer,Dinger,&Mattick,2009)(Supplementary file S1).Of those 28 transcripts with a predicted ORF,only nine(32.1%)had identifiable PFAM domains(Fig.3B table inset).
Of these nine transcripts,one(contig 7074),was identified as sharing homology with the SOX transcription factor family,specifically Sox14(evalue=0.0,381/428,89%identical amino acids,without gaps,from Scylla paramamosain)and SoxC(e-value=0.0,339/433,78%identical amino acids,without gaps,from Macrobrachium nipponese).The C.quadricarinatus homologue possesses the High Mobility Group box(HMG)protein domain(residue positions 48-116 aa),which defines the sox gene family and is essential for DNA binding(Stros,Launholt,&Grasser,2007)(Fig.3C).Our phylogenetic analyses of the putative C.quadricarinatus Sox14,with a range of other annotated sox factors,indicate clear clustering of the transcript within the invertebrate clade,sharing closest homology with other crustacean species.
In vertebrates there are estimated to be over 20 sox genes,involved in a myriad of developmental processes(Stros et al.,2007).Many SOX-domain containing genes have also been reported in the invertebrates,with some known to have important developmental functions in invertebrate model systems(Phochanukul&Russell,2010).In the contextof retinaldevelopment and regeneration,a SoxB transcription factor has been shown to have an eye-determining role in the planarian,S.mediterrane(Lapan&Reddien,2012).Indeed,the authors suggests that the SoxB family encodes a group of genes with an ancestral role in eye biology.SoxB genesin Drosophila,specifically sox-neuro and fish-hook,are expressed in the eye disc(Mukherjee,Shan,Mutsuddi,Ma,&Nambu,2000).Whilst in the vertebrates,sox2 has been shown to be involved in the maintenance of neural progenitor cells and differentiation of retinal ganglion cells(Matsushima,Heavner,&Pevny,2011),while sox15 is involved in the regeneration of skeletal muscle tissue(Lee et al.,2004).Given the lack of functional assessment,we suggest this up-regulated retinal transcription factor,as a C.quadricarinatus sox14 homologue,which warrants further functional investigation in the context of the regenerative capacity of the crustacean eye.
4.Conclusions
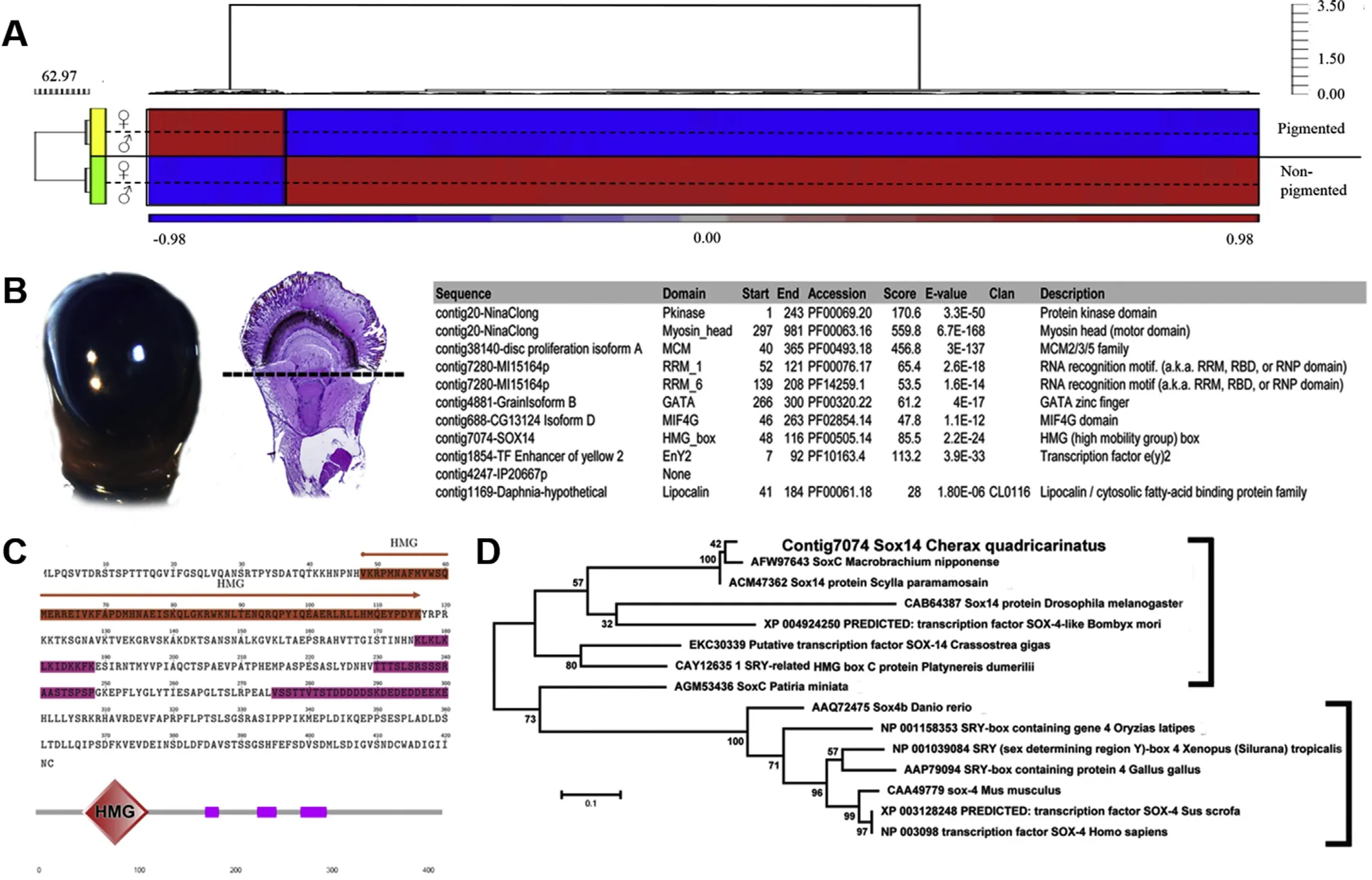
Fig.3.Differential expression of transcripts in pigmented and non-pigmented eye regions,highlighting the predicted sox14 homologue,up-regulated in the pigmented region.A)Heat map of transcripts differentially expressed between the pigmented(retinal)and non-pigmented regions of the C.quadricarinatus eye.Red=up-regulation,blue=down-regulation(normalized scale between 1 and-1).The differentially expressed transcripts(at the 2-fold level)are clustered into two distinct populations:up-regulated in the non-pigmented region(bottom,1163 transcripts)and up-regulated in the pigmented region(top,162 transcripts).B)Sections of the eye:left panel shows the retinal section,while right shows a histological overview of the entire eye with a dashed line showing the separation of retinal(pigmented)and muscular-cerebral(non-pigmented)ocular tissue.The right inset shows a list of genes identified from retinal(pigmented)tissue with PFAM domains(found in 9 of the 162 transcripts up-regulated in the retinal section of the eye).C)ORF of the predicted Sox14 homologue in C.quadricarinatus.Orange highlights the HMG domain,pink the variable domains of low complexity,as predicted by SMART(Schultz et al.,1998).D)Phylogenetic relationships of C.quadricarinatus sox14 transcript and other sox gene family members.Invertebrate species are denoted by the right square bracket.(For interpretation of the references to colour in this figure legend,the reader is referred to the Web version of this article.)
This work is,to our knowledge,the first to provide a transcriptomic description of the ocular tissue in a crustacean species,highlighting notable conservation with the genes functioning in ocular development in Drosophila.Interestingly,re-differentiation from eye to antennae observed in this study is also in keeping with other arthropod mechanisms,indicating a conserved evolution within Arthropoda.We then consider the regenerative capacity of the Crustacea in the context of ocular regeneration,describing the phenotypic progression of retinal regeneration and evaluating the putative modes through which this may be regulated.These modes being(1)a regenerative mechanism similar to that described for limb regeneration,initiated through local progenitor cells,putatively present in the adjacent non-pigmented ocular tissue or(2)through the re-differentiation of the remaining retinal cells themselves,achieved by reconfiguring the molecular environment present during primary development(referred to as the “developmental niche”),similar to the mechanism that has been described in teleost vertebrates,with support from the Planaria.We have identified a putative Sox14 transcription factor n C.quadricarinatus,which warrants further functional investigation in this context.These investigations could validate the overall mode through which regeneration is mediated,confirming if it is indeed through the re-differentiation of the retinal cells;RNAi mediated knock-down of sox14 in a(retina)ablated eye and subsequent comparison of regenerative success would prove a powerful tool in tackling this question.
In addition,this work provides a ready example of the significance of non-model species in gaining a more representative understanding of key developmental processes,which are often limited to a handful of model species.We advocate for the application of aquaculture species,such as C.quadricarinatus,as optimal models to study these processes,such as that described here.The growing availability and relative affordability of transcriptomic analyses offer the opportunity for aquaculture species research to achieve further reach and impact than that directly relating to aquaculture alone,an opportunity not to be missed.
Acknowledgments
This work was supported by funding from the BrightFocus Foundation,and a Ramaciotti Establishment Grant to AWH.
Appendix A.Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.aaf.2018.04.001.
 Aquaculture and Fisheries2019年1期
Aquaculture and Fisheries2019年1期
- Aquaculture and Fisheries的其它文章
- Editorial:Aquaculture genomics
- Uncovering the immunological repertoire of the carpet shell clam Ruditapes decussatus through a transcriptomic-based approach
- SuperSAGE digital expression analysis of differential growth rate in a European sea bass population
- Sex-biased gene discovery from the gonadal transcriptomes of the large yellow croaker(Larimichthys crocea)
- Genome-wide association study identifies loci for body shape in the large yellow croaker(Larimichthys crocea)
