A genetic variant in pseudogene E2F3P1contributes to prognosis of hepatocellular carcinoma
Yun Pn,Chongqi Sun,Ming Hung,Yo Liu,Fuzhn Qi,Li Liu,Jun Wn,Jibin Liu, Kipng Xi,Hongxi M,Zhibin Hu,Hongbing Shn,?
aDepartment of Epidemiology and Biostatistics,Jiangsu Key Laboratory of Cancer Biomarkers,Prevention and Treatment, Cancer Center,School of Public Health,Nanjing Medical University,Nanjing,Jiangsu 211166,China;
bDepartment of Oncology,Huai′an First People′s Hospital,Nanjing Medical University,Huai′an,Jiangsu 223300,China;
cPathology Center and Department of Pathology,Soochow University,Suzhou,Jiangsu 215123,China;
dDepartment of Hepatopancreatobiliary Surgery,Huai′an First People′s Hospital,Nanjing Medical University,Huai′an,
Jiangsu 223300,China;
eDigestive Endoscopy Center,the First Affiliated Hospital of Nanjing Medical University,Nanjing,Jiangsu 210029,China;
fDepartment of Hepatobiliary Surgery,Nantong Tumor Hospital,Nantong,Jiangsu 226361,China.
A genetic variant in pseudogene E2F3P1contributes to prognosis of hepatocellular carcinoma
Yun Pana,△,Chongqi Suna,△,Mingde Huangb,△,Yao Liuc,Fuzhen Qid,Li Liue,Juan Wena,Jibin Liuf, Kaipeng Xiea,Hongxia Maa,Zhibin Hua,Hongbing Shena,?
aDepartment of Epidemiology and Biostatistics,Jiangsu Key Laboratory of Cancer Biomarkers,Prevention and Treatment, Cancer Center,School of Public Health,Nanjing Medical University,Nanjing,Jiangsu 211166,China;
bDepartment of Oncology,Huai′an First People′s Hospital,Nanjing Medical University,Huai′an,Jiangsu 223300,China;
cPathology Center and Department of Pathology,Soochow University,Suzhou,Jiangsu 215123,China;
dDepartment of Hepatopancreatobiliary Surgery,Huai′an First People′s Hospital,Nanjing Medical University,Huai′an,
Jiangsu 223300,China;
eDigestive Endoscopy Center,the First Affiliated Hospital of Nanjing Medical University,Nanjing,Jiangsu 210029,China;
fDepartment of Hepatobiliary Surgery,Nantong Tumor Hospital,Nantong,Jiangsu 226361,China.
Certain pseudogenes may regulate their protein-coding cousins by competing for miRNAs and play an active biological role in cancer.However,few studies have focused on the association of genetic variations in pseudogenes with cancer prognosis.We selected six potentially functional single nucleotide polymorphisms(SNPs)in cancerrelated pseudogenes,and performed a case-only study to assess the association between those SNPs and the prognosis of hepatocellular carcinoma(HCC)in 331 HBV-positive HCC patients without surgical treatment.Log-rank test and Cox proportional hazard models were used for survival analysis.We found that the A allele of rs9909601 in E2F3P1 was significantly associated with a better prognosis compared with the G allele[adjusted hazard ratio(HR)=0.69, 95%confidence interval(CI)=0.56-0.86,P=0.001].Additionally,this protective effect was more predominant for patients without chemotherapy and transcatheter hepatic arterial chemoembolization(TACE)treatment.Interestingly, we also detected a statistically significant multiplicative interaction between genotypes of rs9909601 and chemotherapy or TACE status on HCC survival(P for multiplicative interaction<0.001).These findings indicate that rs9909601 in the pseudogene E2F3P1 may be a genetic marker for HCC prognosis in Chinese.
pseudogene,E2F3P1,SNP,hepatocellular carcinoma(HCC),prognosis
INTRODUCTION
Liver cancer is the fifth most common cancer worldwide and the second most frequent cause of cancer mortality with over 748,300 new cases every year,half of which are in China[1,2].Hepatocellular carcinoma (HCC)is the most common type of liver cancer[3]. Although surgical resection and liver transplantation are regarded as the best treatment for a curative prognosis of early-stage HCC[4],about 85%patients are not suitable for surgery due to locally advanced tumor or distant metastasis[5].Discovery and application of biomarkers that incorporate with traditional cancer staging improve patient care.Thus,substantial efforts have been made to identify biomarkers as prognostic factors for improving therapeutic effect and prognosis prediction.
Pseudogenes are structurally similar to genes that encode functional proteins,but unable to encode fully functional proteins in most cases.Thus,pseudogenes have long been considered as nonfunctional sequences of genomic DNA.However,emerging evidence suggests that pseudogenes may harbor the potential to regulate the expression of their ancestral protein-coding genes by serving as a source of small interfering RNAs(siRNAs),antisense transcripts,microRNA (miRNA)binding sites,or competing mRNAs[6-8]. Furthermore,pseudogenes regulate tumor suppressors and oncogenes by acting as microRNA decoys[9-14].To date,several studies have reported the association between pseudogene expression and multiple cancer risk.Ishiguro et al.reported that two pseudogenes (NANOG1 and NANOGP8)were differentially expressed in colon cancer cells,and their expression might contribute to the proliferation of colon cancer cells[15].Similar results were found for the association of POU5F1P1 expression with prostatic carcinoma[16],PTENP1 expression with melanoma[17],and NANOGP8 expression with gastric cancer[18].However,little is known about pseudogenes and cancer prognosis.
Functional polymorphisms in pseudogenes,such as single nucleotide polymorphisms(SNPs)influencing miRNAs binding,may affect the expression or function of the proteins[19,20].Thus,we speculate that potentially functional polymorphisms in pseudogenes may affect the expression of pseudogenes or its original protein-coding genes by influencing miRNA binding affinity,and thus play a role in the development and progression of human cancer.In this study,we examined the associations between six genetic variants of pseudogenes and prognosis of 331 patients with intermediate or advanced HCC in Chinese.
SUBJECTS AND METHODS
Subjects
The protocol was approved by the local institutional review board at the authors′affiliated institution. Written informed consent was obtained from every subject.The enrollments of subjects were described previously[21,22].For constructing a relatively homogenous population,our current study was restricted to HCC patients without surgery in intermediate stage(B) or advanced stage(C)according to the Barcelona Clinic Liver Cancer(BCLC)staging system[23,24].We recruited 414 intermediate or advanced HCC patients from Nantong Tumor Hospital and the First Affiliated Hospital of Nanjing Medical University,Nanjing, Jiangsu,China.All patients were followed up prospectively every three months from the time of enrollment by personal or family contacts until death or last time of follow-up.As a result,a total of 331 intermediate or advanced HCC patients who had completed follow-ups and clinical information were enrolled in our study with a response rate of 80.0%.The maximum follow-up time (MFT)for the 331 patients involved in the present study was 60.7 months(last follow-up in January 2013)and the median survival time(MST)was 14.5 months.
Serological tests
HBsAg,anti-HBs,anti-HBc and anti-HCV were detected by the enzyme-linked immunosorbent assay (Kehua Bio-engineering Co.,Ltd.,Shanghai,China) following the manufacturer′s instructions as described previously[21].
Selection and genotyping of SNPs
We included eight key cancer-related pseudogenes from a review conducted by Poliseno et al.[8](Supplemental Table 1available online).By blasting the identical sequence between pseudogenes and their parental genes,we found 31 common SNPs located in pseudogenes,with at least 50 bps flanking regions identical to their parental genes.Then,we searched miRBase (http://www.mirbase.org/)and Patrocles(http://www. patrocles.org/)to find whether the allelic change of 31 SNPs may influence miRNAs binding.Eventually,we selected six potentially functional SNPs that might affect miRNA binding(rs11682718 in DNMT3AP1,rs1838149 and rs9909601 in E2F3P1,rs2004079 in NANOGP8, rs9889937 in FOXO3B,and rs6913881 in KRASP1).
Genomic DNA was extracted from a leukocyte pellet by traditional proteinase K digestion,phenolchloroform extraction and ethanol precipitation.All SNPs were genotyped using the TaqMan allelic discri-mination assay on a 7900 system(Applied Biosystems, Carlsbad,CA,USA).The primers and probes for the six SNPs are showninSupplemental Table 2(available online).Two blank(water)controls in each 384-well plate were performed for quality control,and more than 5%samples were randomly selected and repeated, yielding a 100%concordance.Thesuccess ratesof genotyping for the six SNPs were all above 95%.
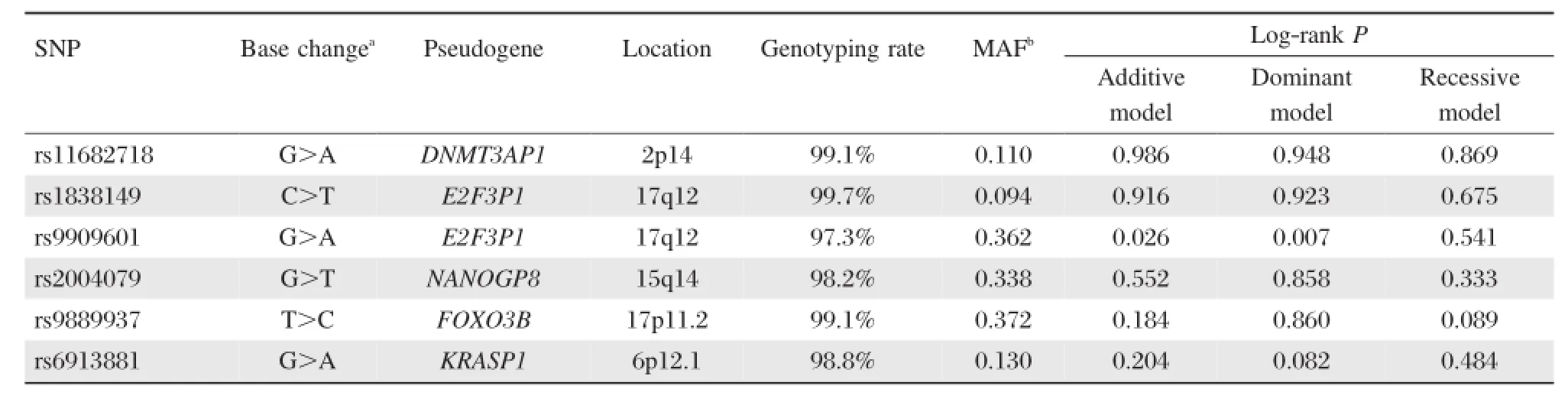
Table 1Genotyping results of survival of HCC patients
Statistical analysis
Mean survival time was presented when the MST could not be calculated.Kaplan-Meier method and log-rank test were performed to compare the survival time in different subgroups categorized by patient characteristics,clinical features and genotypes.Univariate and multivariable Cox proportional hazard regression analyses were performed to estimate the crude or adjusted hazard ratio(HR)and their 95%confidence intervals(CI),with adjustment of age,gender,smoking status,drinking status,BCLC stage,and chemotherapy or transcatheter hepatic arterial chemoembolization (TACE)status.Cox stepwise regression model was also conducted to determine predictive factors of HCC prognosis,with a significance level of 0.050 for entering and 0.051 for removing the respective explanatory variables.The Chi-square-based Q test was applied to test the heterogeneity of associations between subgroups. Analyses were carried out using Statistical Analysis System software(version 9.1.3;SAS Institute,Cary, NC,USA).All tests were two-sided and the criterion of statistical significance was set at P<0.05.
RESULTS
Demographic and baseline characteristics of the study subjects
The demographic characteristics and clinical information of the 331 HCC patients in stage B or C included in the study were described previously[22]. Totally,258 of them died from HCC,and two died from other causes during up to 60.7 months of follow-up.For disease-specific survival analysis,the latter were considered as censored data in the analyses. Drinking and chemotherapy or TACE status was significantly associated with survival time(log-rank P=0.006 and<0.001 for drinking status and chemotherapy or TACE status,respectively).Notably, compared to those who received neither chemotherapy nor TACE therapy(MST=3.4 months),patients with chemotherapy or TACE therapy(MST=16.8 months) had a 61%significantly decreased risk of death(HR= 0.39;95%CI=0.29-0.51).
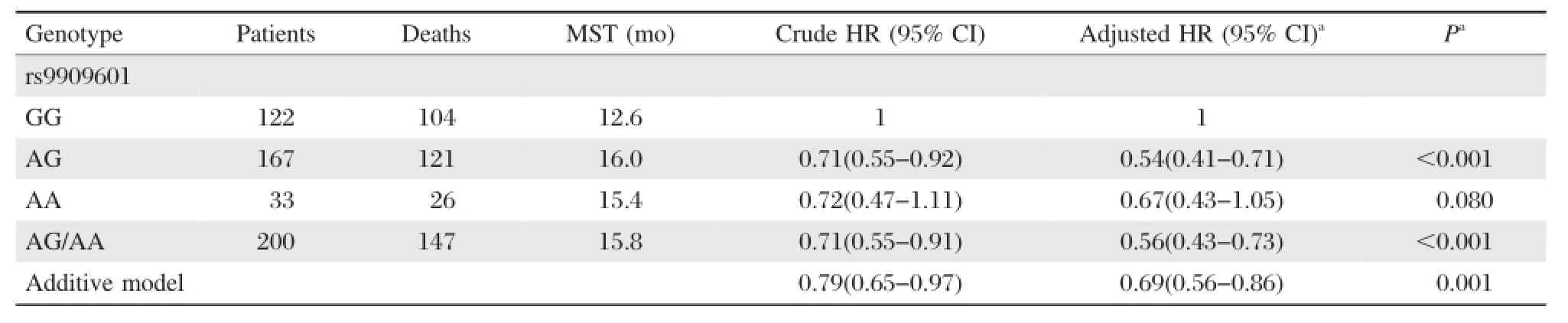
Table 2Genotypes ofE2F3P1rs9909601 and survival of HCC patients
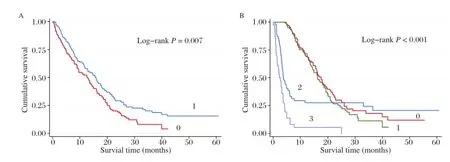
Fig.1Kaplan-Meier plots of survival byE2F3P1rs9909601 genotypes in the survival of hepatocellular carcinoma(HCC)patients. A:E2F3P1 rs9909601 genotypes and HCC survival(log-rank P=0.007 for AG/AA vs.GG)in a dominant model.0,patients with common genotype GG; 1,those with variant genotypes(AG/AA).B:Kaplan-Meier plots of survival by the combination of rs9909601 genotypes and chemotherapy or transcatheter hepatic arterial chemoembolization(TACE)therapy status in HCC-specific survival(log-rank P<0.001).0,patients with variant genotypes(AG/AA)and receiving chemotherapy or TACE therapy;1,those with common genotype(GG)and receiving chemotherapy or TACE therapy;2,those with variant genotypes(AG/AA)and without chemotherapy and TACE therapy;3,those with GG genotype and without chemotherapy and TACE therapy.
Effects of polymorphisms in pseudogenes on HCC survival
Kaplan-Meier method and log-rank test were performed to examine the associations of the six SNPs with HCC survival in different genetic models(additive model,dominant model and recessive model). As shown inTable 1,the difference between the survival time of HCC patients and variant genotypes of rs9909601 located in E2F3P1 was statistically significant(log-rank test:P=0.007 in the dominant model and P=0.026 in the additive model,respectively). Patients carrying rs9909601 AG/AA genotypes survived significantly longer time(MST=15.8 months) than those carrying rs9909601GG genotypes(MST= 12.6 months;Fig.1A).Furthermore,multivariable Cox regression analysis showed that rs9909601 remained as a significant prognostic marker for HCC (Table 2).After adjusting for age,gender,smoking status,drinking status,BCLC stage,and chemotherapy or TACE status,variant genotypes of rs9909601were significantly associated with favorable prognosis of HCC(additive model:adjusted HR=0.69,95%CI= 0.56-0.86,P=0.001;dominant model:adjusted HR =0.56,95%CI=0.43-0.73,P<0.001).
Stepwise Cox regression analysis on HCC survival
Then,we performed stepwise Cox proportional hazard analysis to estimate the effects of demographic characteristics,clinical features,and E2F3P1 rs9909601 on HCC survival.As shown inTable 3,four variables (age,drinking status,chemotherapy or TACE status,and E2F3P1 rs9909601)were selected into the final regression model.Furthermore,when gender,smoking status and BCLC stage were included in the final model, the E2F3P1 rs9909601 still remained as an independent protective factor for HCC survival(HR=0.56,95% CI=0.43-0.72,P<0.001).
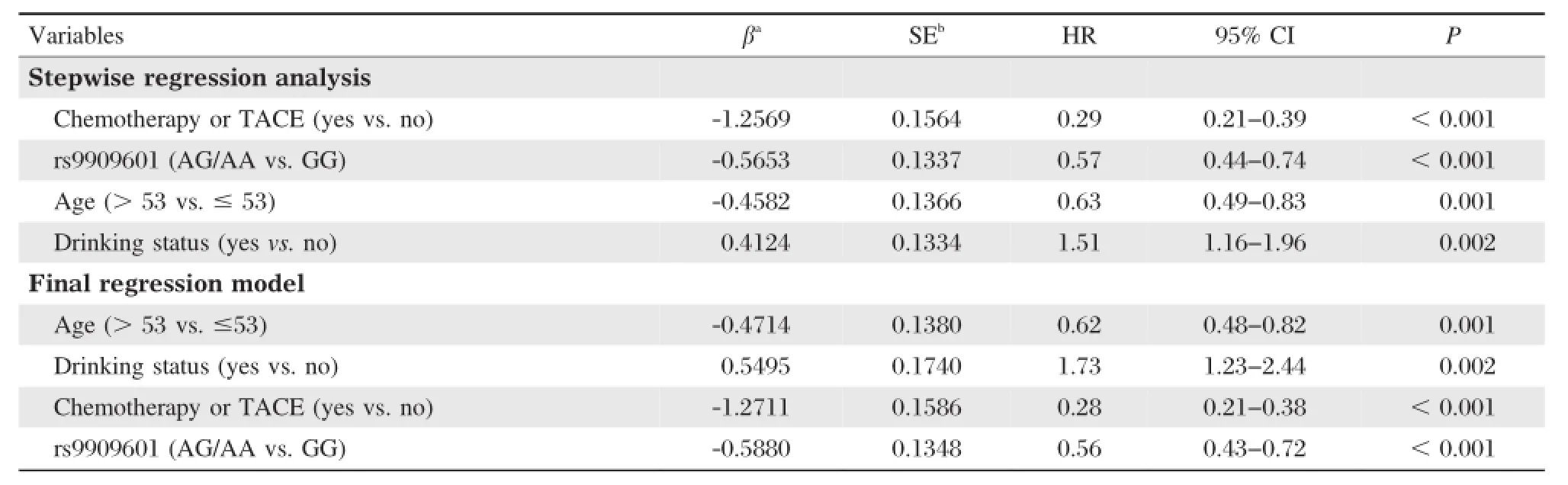
Table 3Multivariable Cox regression analysis on survival of HCC patients
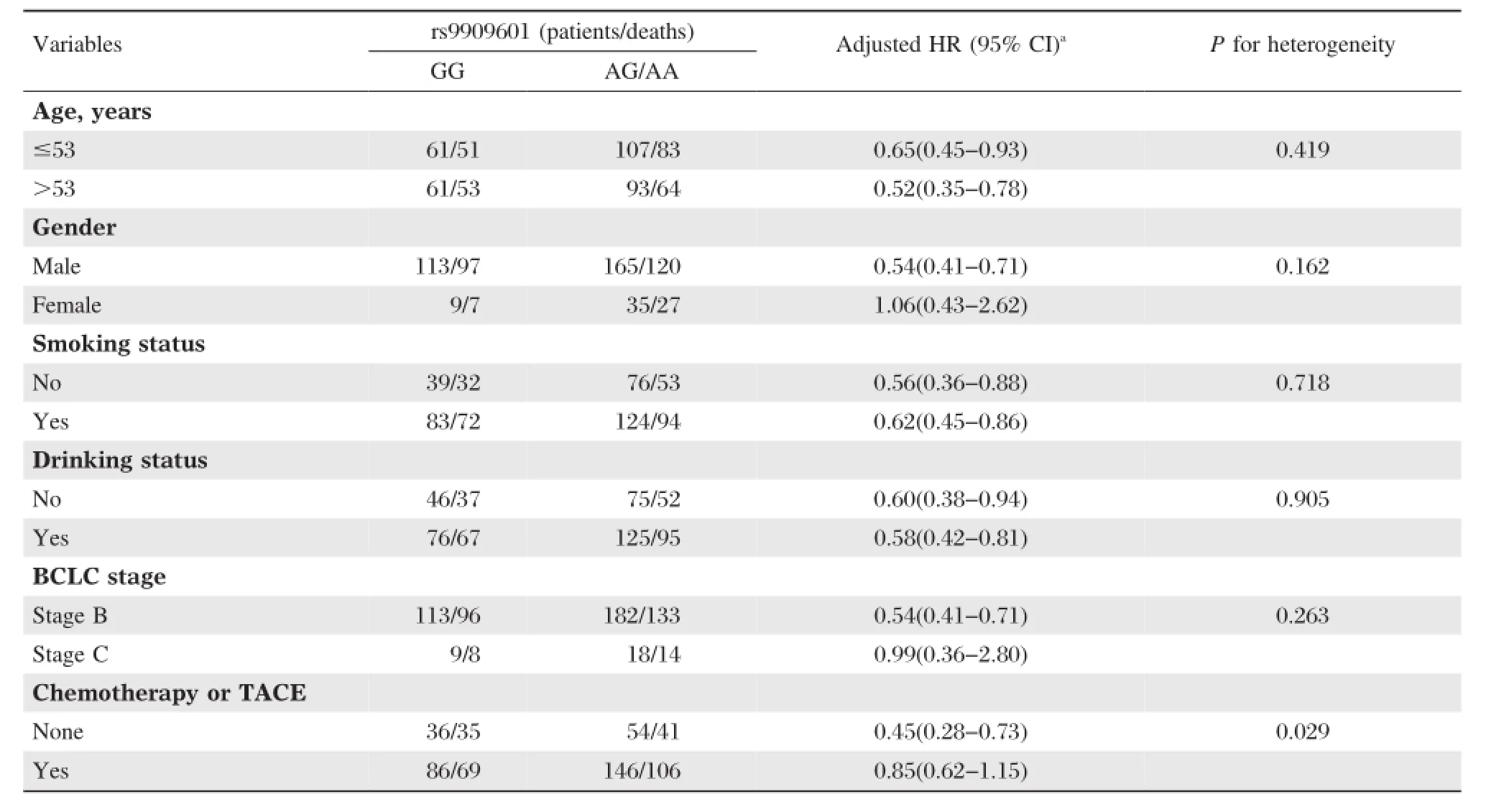
Table 4Stratification analysis of rs9909601 genotypes associated with survival of HCC patients
Stratification and interaction analysis
The associations between E2F3P1 rs9909601 and HCC survival were further investigated by stratification of age,gender,smoking status,drinking status,BCLC stage,and chemotherapy or TACE status.As shown inTable 4,we found that the protective effect of rs9909601 variant genotypes was more prominent in patients without chemotherapy and TACE(adjusted HR=0.45,95%CI=0.28-0.73)than those with chemotherapy or TACE therapy(adjusted HR=0.85, 95%CI=0.62-1.15,P=0.029 for heterogeneity test). Therefore,a gene-chemotherapy or TACE status interaction analysis was carried out,and a statistically significant multiplicative interaction was observed (P for multiplicative interaction<0.001,Fig.1B). Compared to subjects with AG/AA genotypes and with chemotherapy or TACE therapy,patients with GG genotype but without chemotherapy or TACE therapy had a significantly increased mortality risk(adjusted HR= 14.98,95%CI=9.20-24.37,P<0.001)(Table 5).
DISCUSSION
In the present study,we investigated the effects of six common SNPs in cancer-related pseudogenes on the survival of advanced HCC patients and demon-strated that E2F3P1 rs9909601 may be an independent biomarker to predict the survival of advanced HCC patients.To the best of our knowledge,this is the first report to evaluate the role of genetic variations of pseudogene in HCC survival.
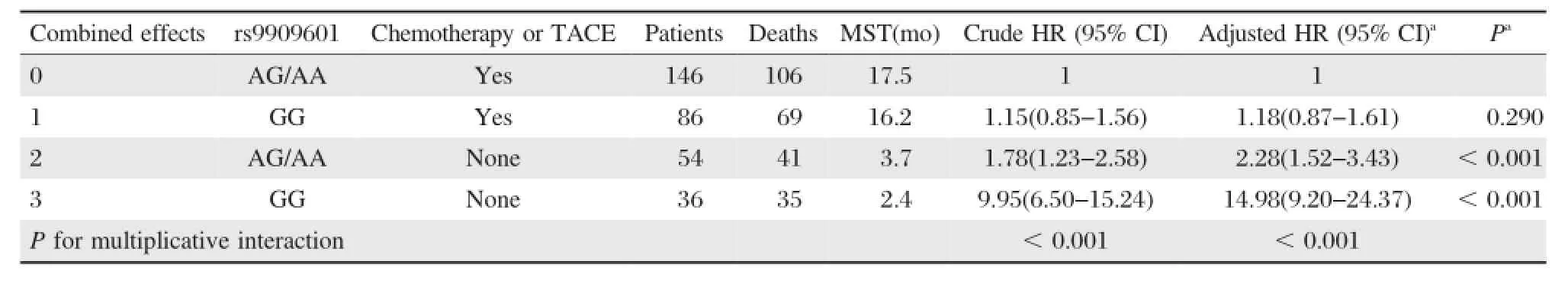
Table 5Interaction between rs9909601 genotypes and chemotherapy or TACE status
E2F was reported to regulate the expression of multiple genes that are important in cell proliferation as a transcription factor[25].Specifically,it plays a critical role in the control of cell cycle[26,27].Recent findings suggested that the expression of E2F3 was regulated by several miRNAs[28]and played a major role in modifying cellular proliferation rate which directly or indirectly affected clinical outcome of many types of tumors, including bladder cancer[29],prostate cancer[30],ovarian cancer[31]and breast cancer[32].E2F3P1 located in chromosome 17 is a pseudogene with sequence similar to E2F3 in chromosome 6.Although they are in different chromosomes,E2F3P1 may regulate E2F3 expression by competing for miRNAs and their expressions are positively correlated[33].Lees et al.have reported that genetic variations in E2F3P1 may influence the miRNAs binding and thus may interrupt subsequent cellular activity,including proliferation and apoptosis[34]. Thus,it is biologically plausible that genetic variations in pseudogenes contribute to cancer risk or prognosis, given that miRNAs may provide linkage between pseudogenes and their parent genes.
By using two web-based prediction tools(miRBase and Patrocles),we found that the wild G allele of E2F3P1 rs9909601 was more inclined to bind miR-24,miR-149,and miR-892b than the variant A allele. Both of the miR-24 and miR-149 have been investigated substantially in cancers.For instance,Han et al.identified that the overexpression of miR-24 was associated with the non-recurrence of hepatocellular carcinoma following liver transplantation which contributed to a better prognosis of HCC[35].Besides,several studies have indicated that the down-regulation of miR-149 has been found in a variety of carcinomas and finally led to a worse patient survival,such as head and neck squamous cell carcinoma[36],colorectal cancer[37]and astrocytoma[38].Thus,E2F3P1 rs9909601 identified in our study may affect the impact of miRNAs regulation on gene expression by influencing the binding affinity of several special miRNAs,and hence play a role in the progression of HCC.Moreover,our analyses indicated a significant interaction between variant genotypes of rs9909601 and therapy status,which provided evidence that the effect of genetic variants on HCC prognosis could be modified by clinical factors.
In conclusion,rs9909601 at E2F3P1 may be a useful biomarker for the prognosis of HCC survival. However,other studies with larger sample size and functional analysis are warranted to verify our finding.
Acknowledgements
We would like to thank Dr.Juncheng Dai for his excellent technical assistance.
REFERENCES
[1] Jemal A,Bray F,Center MM,Ferlay J,Ward E,Forman D.Global cancer statistics.CA Cancer J Clin 2011;61: 69-90.
[2] Parkin DM,Bray F,Ferlay J,Pisani P.Global cancer statistics,2002.CA Cancer J Clin 2005;55:74-108.
[3] Lau WY,Lai EC.Hepatocellular carcinoma:current management and recent advances.Hepatobiliary Pancreat Dis Int 2008;7:237-57.
[4] Liang KH,Lin CC,Yeh CT.GALNT14 SNP as a potential predictor of response to combination chemotherapy using 5-FU,mitoxantrone and cisplatin in advanced HCC.Pharmacogenomics 2011;12:1061-73.
[5] LIovet JM,Bruix J,Gores GJ.Surgical resection versus transplantation for early hepatocellular carcinoma:clues for the best strategy.Hepatology 2000;31:1019-21.
[6] Han YJ,Ma SF,Yourek G,Park YD,Garcia JG.A transcribed pseudogene of MYLK promotes cell proliferation. FASEB J 2011;25:2305-12.
[7] Hawkins PG,Morris KV.Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5.Transcription 2010;1:165-75.
[8] Poliseno L,Salmena L,Zhang J,Carver B,Haveman WJ, Pandolfi PP.A coding-independent function of gene and pseudogene mRNAs regulates tumour biology.Nature 2010;465:1033-8.
[9] Salmena L,Poliseno L,Tay Y,Kats L,Pandolfi PP.A ceRNA hypothesis:the Rosetta Stone of a hidden RNA language?Cell 2011;146:353-8.
[10]Poliseno L.Pseudogenes:newly discovered players in human cancer.Sci Signal 2012;5:re5.
[11]Kalyana-Sundaram S,Kumar-Sinha C,Shankar S, Robinson DR,Wu YM,Cao X,et al.Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 2012;149:1622-34.
[12]Tam OH,Aravin AA,Stein P,Girard A,Murchison EP, Cheloufi S,et al.Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008;453:534-8.
[13]Zhou BS,Beidler DR,Cheng YC.Identification of antisense RNA transcripts from a human DNA topoisomerase I pseudogene.Cancer Res 1992;52:4280-5.
[14]Kandouz M,Bier A,Carystinos GD,Alaoui-Jamali MA, Batist G.Connexin43 pseudogene is expressed in tumor cells and inhibits growth.Oncogene 2004;23:4763-70.
[15]Ishiguro T,Sato A,Ohata H,Sakai H,Nakagama H, Okamoto K.Differential expression of nanog1 and nanogp8 in colon cancer cells.Biochem Biophys Res Commun 2012;418:199-204.
[16]Kastler S,Honold L,Luedeke M,Kuefer R,Mo¨ller P, Hoegel J,et al.POU5F1P1,a putative cancer susceptibility gene,is overexpressed in prostatic carcinoma.Prostate 2010;70:666-74.
[17]Poliseno L,Haimovic A,Christos PJ,Vega Y Saenz de Miera EC,Shapiro R,Pavlick A,et al.Deletion of PTENP1 pseudogene in human melanoma.J Invest Dermatol 2011;131:2497-500.
[18]Zhang J,Wang X,Chen B,Xiao Z,Li W,Lu Y,et al.The human pluripotency gene NANOG/NANOGP8 is expressed in gastric cancer and associated with tumor development.Oncol Lett 2010;1:457-63.
[19]Diskin SJ,Capasso M,Schnepp RW,Cole KA,Attiyeh EF,Hou C,et al.Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma.Nat Genet 2012;44:1126-30.
[20]Ryan BM,Robles AI,Harris CC.Genetic variation in microRNA networks:the implications for cancer research. Nat Rev Cancer 2010;10:389-402.
[21]Hu L,Zhai X,Liu J,Chu M,Pan S,Jiang J,et al.Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development.Hepatology 2012;55:1426-31.
[22]Xie K,Liu J,Zhu L,Liu Y,Pan Y,Juan Wen,et al.A potentially functional polymorphism in the promoter region of let-7 family is associated with survival of hepatocellular carcinoma.Cancer Epidemiol 2013;37:998-1002.
[23]Gomez-Rodrguez R,Romero-Gutierrez M,Artaza-Varasa T,Gonza′lez-Frutos C,Ciampi-Dopazo JJ,de-la-Cruz-Perez G,et al.The value of the Barcelona Clinic Liver Cancer and alpha-fetoprotein in the prognosis of hepatocellular carcinoma.Rev Esp Enferm Dig 2012;104:298-304.
[24]Forner A,Llovet JM,Bruix J.Hepatocellular carcinoma. Lancet 2012;379:1245-55.
[25]Nevins JR.E2F:a link between the Rb tumor suppressor protein and viral oncoproteins.Science 1992;258:424-9.
[26]Wong JV,Dong P,Nevins JR,Mathey-Prevot B,You L. Network calisthenics:control of E2F dynamics in cell cycle entry.Cell Cycle 2011;10:3086-94.
[27]Kong LJ,Chang JT,Bild AH,Nevins JR.Compensation and specificity of function within the E2F family. Oncogene 2007;26:321-7.
[28]Li W,Ni GX,Zhang P,Zhang ZX,Li W,Wu Q. Characterization of E2F3a function in HepG2 liver cancer cells.J Cell Biochem 2010;111:1244-51.
[29]Huang L,Luo J,Cai Q,Pan Q,Zeng H,Guo Z,et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3.Int J Cancer 2011;128:1758-69.
[30]Foster CS,Falconer A,Dodson AR,Norman AR,Dennis N,Fletcher A,et al.Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome.Oncogene 2004;23:5871-9.
[31]Reimer D,Hubalek M,Riedle S,Skvortsov S,Erdel M, Concin N,et al.E2F3a is critically involved in epidermal growth factor receptor-directed proliferation in ovarian cancer.Cancer Res 2010;70:4613-23.
[32]Shi Z,Derow CK,Zhang B.Co-expression module analysis reveals biological processes,genomic gain,and regulatory mechanisms associated with breast cancer progression.BMC Syst Biol 2010;4:74.
[33]Echols N,Harrison P,Balasubramanian S,Luscombe NM,Bertone P,Zhang Z,et al.Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes,comparing genes and pseudogenes.Nucleic Acids Res 2002;30:2515-23.
[34]Lees JA,Saito M,Vidal M,Valentine M,Look T,Harlow E,et al.The retinoblastoma protein binds to a family of E2F transcription factors.Mol Cell Biol 1993;13:7813-25.
[35]Han ZB,Zhong L,Teng MJ,Fan JW,Tang HM,Wu JY, et al.Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol 2012;6:445-57.
[36]Tu HF,Liu CJ,Chang CL,Wang PW,Kao SY,Yang CC, et al.The association between genetic polymorphism and the processing efficiency of miR-149 affects the prognosis of patients with head and neck squamous cell carcinoma. PLoS One 2012;7:e51606.
[37]Wang F,Ma YL,Zhang P,Shen TY,Shi CZ,Yang YZ, et al.SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer.J Pathol 2013;229:12-24.
[38]Li D,Chen P,Li XY,Zhang LY,Xiong W,Zhou M,et al. Grade-specific expression profiles of miRNAs/mRNAs and docking study in human grade I-III astrocytomas. OMICS 2011;15:673-82.
Received 16 March 2014,Accepted 31 March 2014,Epub 27 April 2014
This work was funded by the National Natural Science Foundation of China(81372606 and 81072344),Project supported by the National Key Basic Research Program Grant(2013CB911400),the project supported by the National Science Foundation for Distinguished Young Scholars of China(81225020),Foundation of Jiangsu Province for Distinguished Young Scholars(BK2012042),Foundation for the Program for New Century Excellent Talents in University(NCET-10-0178),the Fok Ying-Tong Education Foundation for Young Teachers in the Higher Education Institutions(122031),Young Tip-top Talents Support Program by the Organization Department of the CPC Central Committee,the Author of National Excellent Doctoral Dissertation(201081),Jiangsu Province Clinical Science and Technology Projects (BL2012008)and the Priority Academic Program for the Development of Jiangsu Higher Education Institutions(Public Health and Preventive Medicine).
△These authors contributed equally to this work.
?Corresponding author:Hongbing Shen,MD,PhD,Department of Epidemiology and Biostatistics,School of Public Health,Nanjing Medical University,818 Tianyuan East Rd.,Nanjing,Jiangsu 211166, China.Tel/Fax:+86-25-8686-8439/+86-25-8686-8439,E-mail:hbshen@ njmu.edu.cn.
The authors reported no conflict of interests.
?2014 by the Journal of Biomedical Research.All rights reserved.
10.7555/JBR.28.20140052
 THE JOURNAL OF BIOMEDICAL RESEARCH2014年3期
THE JOURNAL OF BIOMEDICAL RESEARCH2014年3期
- THE JOURNAL OF BIOMEDICAL RESEARCH的其它文章
- Analytical characteristics of a qPCR-based molecular diagnostic assay-conceptual considerations for laboratory personnel
- Drainage vs.non-drainage after cholecystectomy for acute cholecystitis:a retrospective study
- Position and complications of pedicle screw insertion with or without image-navigation techniques in the thoracolumbar spine: a meta-analysis of comparative studies
- Correlation of obstructive sleep apnea hypopnea syndrome with metabolic syndrome in snorers
- Class A scavenger receptor activation inhibits endoplasmic reticulum stress-induced autophagy in macrophage
- Expression of human hepatic lipase negatively impacts apolipoprotein A-I production in primary hepatocytes from Lipc-null mice
